Gene Information
|
Gene Name
|
FURIN |
|
Gene ID
|
5045
|
|
Gene Full Name
|
furin, paired basic amino acid cleaving enzyme |
|
Gene Alias
|
FUR|PACE|PCSK3|SPC1 |
|
Transcripts
|
ENSG00000140564
|
|
Virus
|
SARS-CoV-2 |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Golgi apparatus;Nucleoplasm;Intracellular and membrane; 
|
|
Membrane Info
|
Enzymes, Plasma proteins, Predicted membrane proteins |
|
Uniport_ID
|
P09958
|
|
HGNC ID
|
HGNC:8568
|
|
OMIM ID
|
136950 |
|
Summary
|
This gene encodes a member of the subtilisin-like proprotein convertase family, which includes proteases that process protein and peptide precursors trafficking through regulated or constitutive branches of the secretory pathway. It encodes a type 1 membrane bound protease that is expressed in many tissues, including neuroendocrine, liver, gut, and brain. The encoded protein undergoes an initial autocatalytic processing event in the ER and then sorts to the trans-Golgi network through endosomes where a second autocatalytic event takes place and the catalytic activity is acquired. Like other members of this convertase family, the product of this gene specifically cleaves substrates at single or paired basic residues. Some of its substrates include proparathyroid hormone, transforming growth factor beta 1 precursor, proalbumin, pro-beta-secretase, membrane type-1 matrix metalloproteinase, beta subunit of pro-nerve growth factor and von Willebrand factor. It is thought to be one of the proteases responsible for the activation of HIV envelope glycoproteins gp160 and gp140, and may play a role in tumor progression. Unlike SARS-CoV and other coronaviruses, the spike protein of SARS-CoV-2 is thought to be uniquely cleaved by this protease. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2020] |
Target gene [FURIN] related to virus gene/protein/region 
Mutation Table: if previous studies reported that target gene was related to virus gene/region/protein, the information in this literature was listed in this section
Target gene [FURIN] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE167075
|
MeRIP-seq;RNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens; Severe acute respiratory syndrome coronavirus 2);Illumina NovaSeq 6000 (Chlorocebus aethiops; Severe acute respiratory syndrome coronavirus 2) |
|
GSE164485
|
scRNA-seq |
36 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE190680
|
RNA-seq |
100 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE207923
|
RNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE152641
|
RNA-seq |
86 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE283771
|
RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE211934
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE163959
|
RNA-seq |
48 |
Illumina NextSeq 500 (Homo sapiens) |
|
C GSE163623
|
Chip-seq |
5 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE240694
|
scRNA-seq |
9 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE166253
|
RNA-seq |
26 |
HiSeq X Ten (Homo sapiens) |
|
GSE153218
|
RNA-seq |
16 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE162323
|
RNA-seq |
42 |
Illumina NovaSeq 6000 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens; Severe acute respiratory syndrome coronavirus 2) |
|
GSE199816
|
RNA-seq |
27 |
DNBSEQ-G400 (Homo sapiens) |
|
E GSE215262
|
RNA-seq |
25 |
Illumina NovaSeq 6000 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE215262 - FURIN expression across samples
|
Volcano Plot
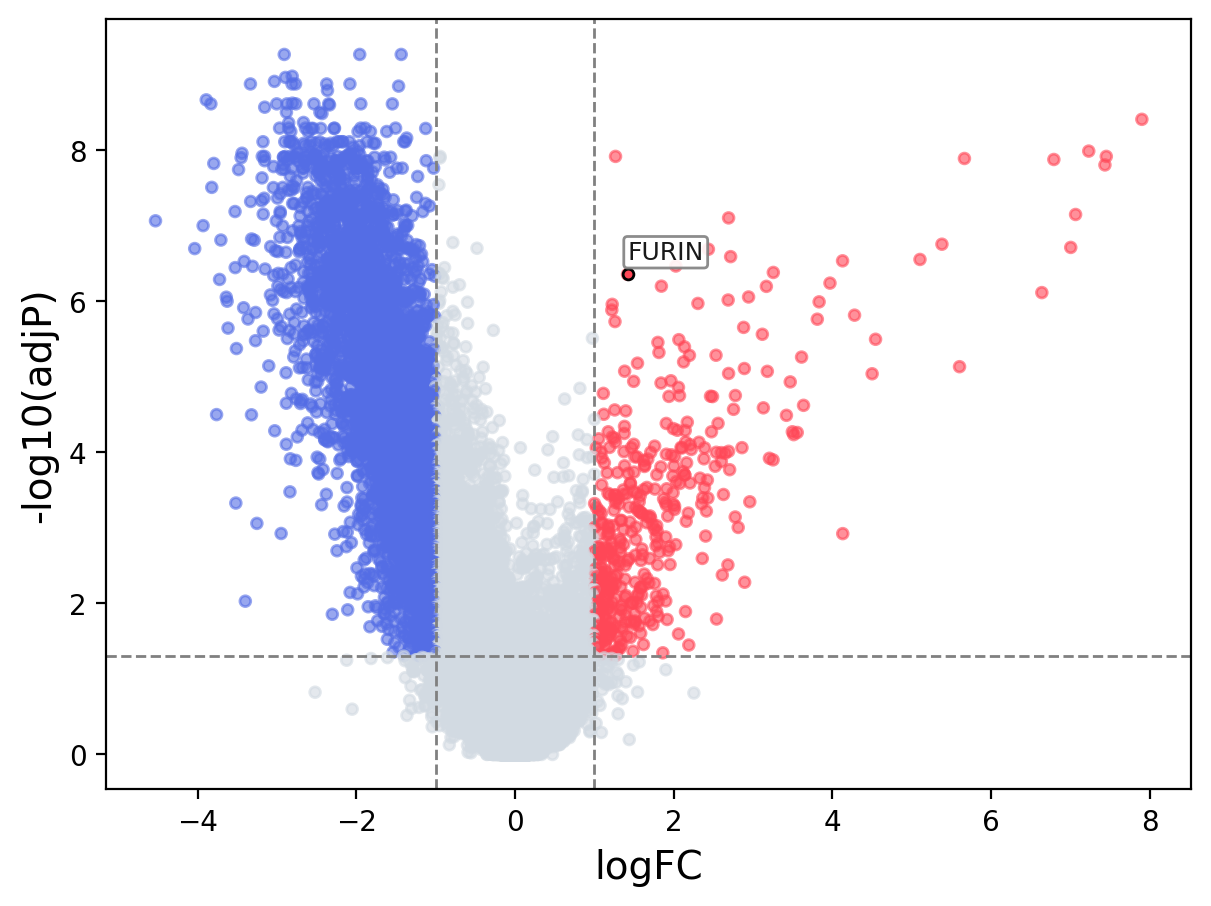
|
Bar Plot
|
|
|
 SARS-CoV-2 Target gene Detail Information
SARS-CoV-2 Target gene Detail Information