Gene Information
|
Gene Name
|
GNAS |
|
Gene ID
|
2778
|
|
Gene Full Name
|
GNAS complex locus |
|
Gene Alias
|
AHO|AIMAH1|C20orf45|GNAS1|GPSA|GSA|GSP|NESP|PITA3|POH|SCG6|SgVI |
|
Transcripts
|
ENSG00000087460
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Plasma membrane, Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Potential drug targets, Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
O95467, P63092, Q5JWF2
|
|
HGNC ID
|
HGNC:4392
|
|
OMIM ID
|
139320 |
|
Summary
|
This locus has a highly complex imprinted expression pattern. It gives rise to maternally, paternally, and biallelically expressed transcripts that are derived from four alternative promoters and 5' exons. Some transcripts contain a differentially methylated region (DMR) at their 5' exons, and this DMR is commonly found in imprinted genes and correlates with transcript expression. An antisense transcript is produced from an overlapping locus on the opposite strand. One of the transcripts produced from this locus, and the antisense transcript, are paternally expressed noncoding RNAs, and may regulate imprinting in this region. In addition, one of the transcripts contains a second overlapping ORF, which encodes a structurally unrelated protein - Alex. Alternative splicing of downstream exons is also observed, which results in different forms of the stimulatory G-protein alpha subunit, a key element of the classical signal transduction pathway linking receptor-ligand interactions with the activation of adenylyl cyclase and a variety of cellular reponses. Multiple transcript variants encoding different isoforms have been found for this gene. Mutations in this gene result in pseudohypoparathyroidism type 1a, pseudohypoparathyroidism type 1b, Albright hereditary osteodystrophy, pseudopseudohypoparathyroidism, McCune-Albright syndrome, progressive osseus heteroplasia, polyostotic fibrous dysplasia of bone, and some pituitary tumors. [provided by RefSeq, Aug 2012] |
Target gene [GNAS] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5008318 |
chr20 |
4 |
2 |
0 |
6 |
View |
Target gene [GNAS] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE139324
|
scRNA-seq |
63 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
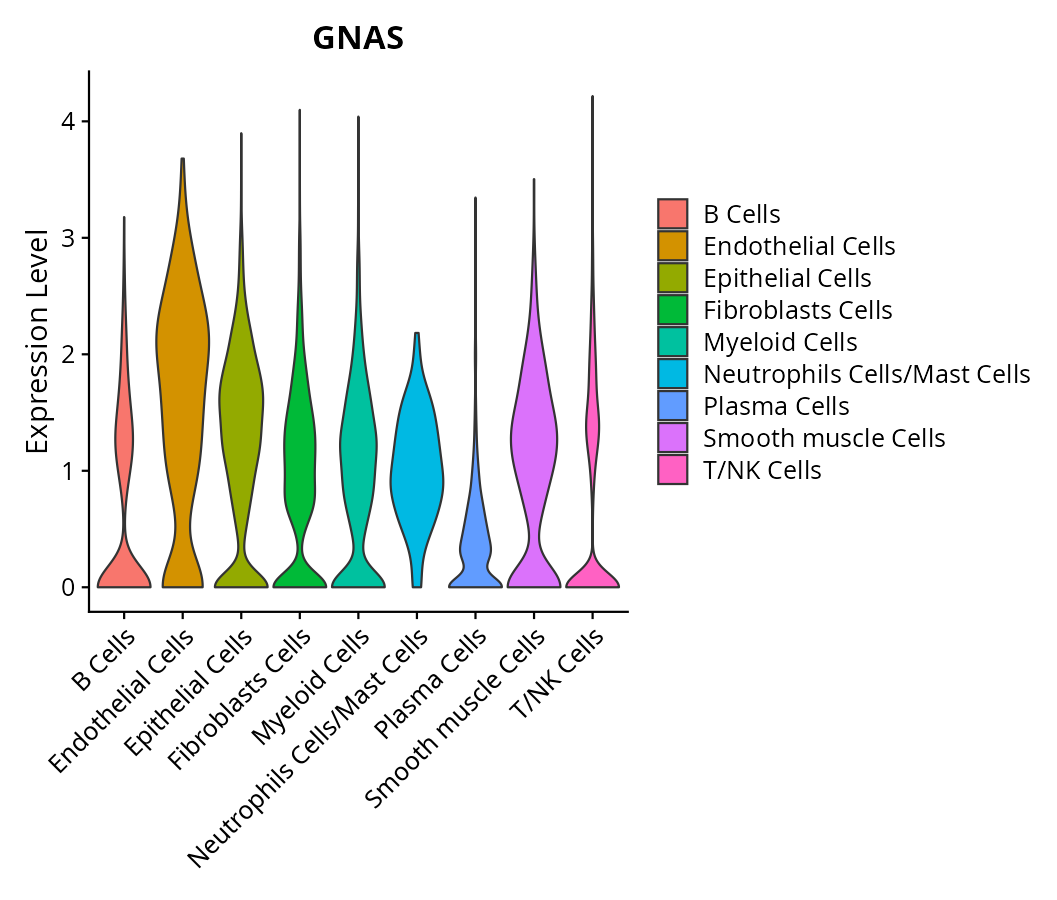
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE197461 - Gene expression in cell subsets
|
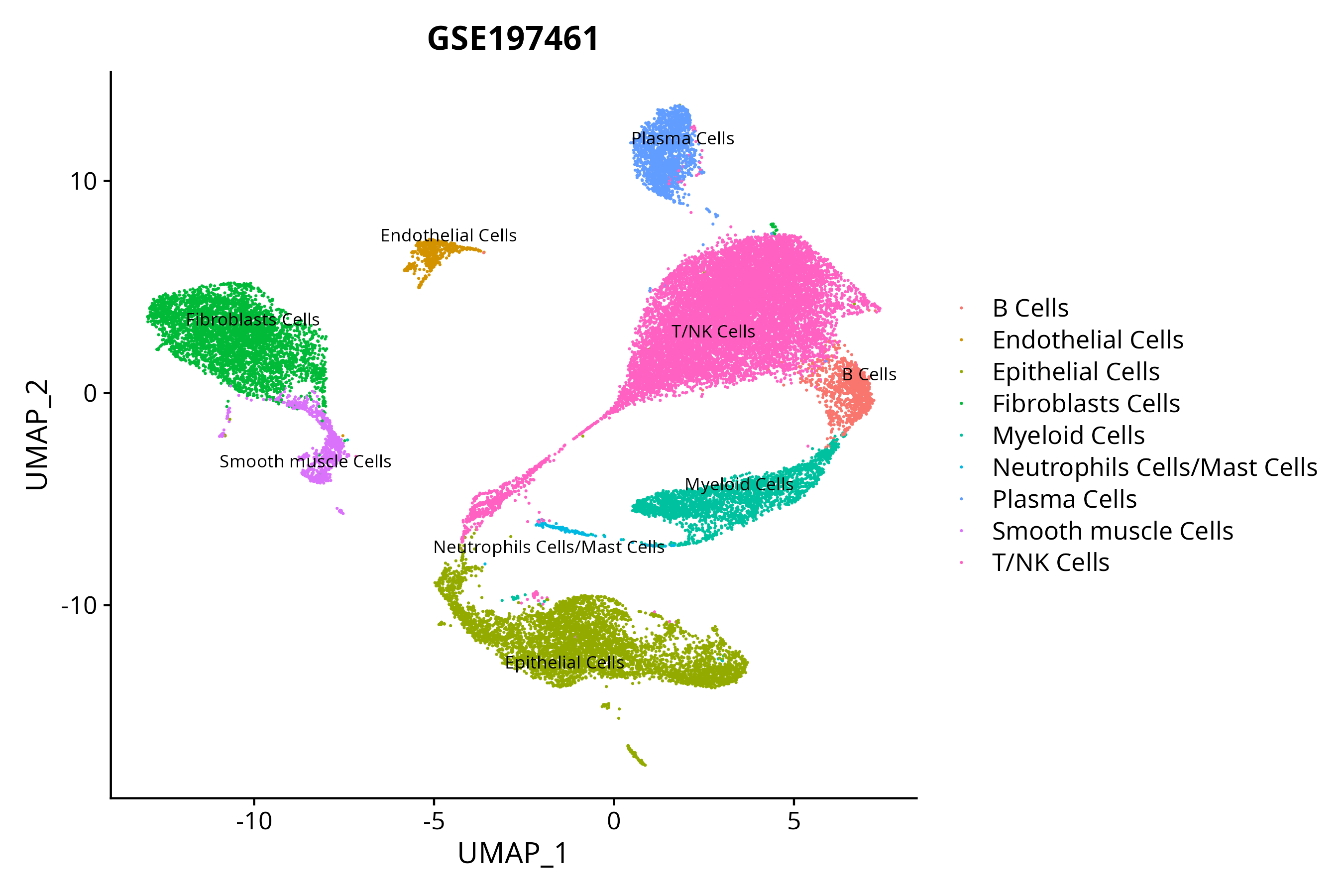
|
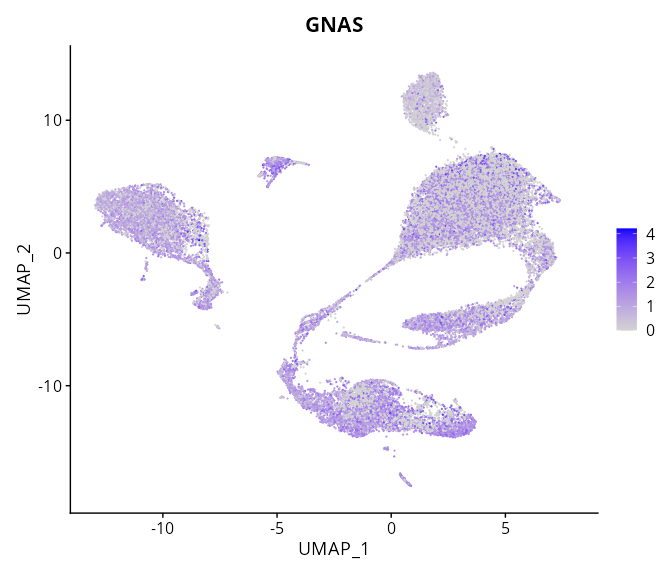
|
|
|
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information