Gene Information
|
Gene Name
|
HLA-B |
|
Gene ID
|
3106
|
|
Gene Full Name
|
major histocompatibility complex, class I, B |
|
Gene Alias
|
AS|B-4901|HLAB |
|
Transcripts
|
ENSG00000234745
|
|
Virus
|
SARS-CoV-2 |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P01889
|
|
HGNC ID
|
HGNC:4932
|
|
OMIM ID
|
142830 |
|
Summary
|
HLA-B belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. Class I molecules play a central role in the immune system by presenting peptides derived from the endoplasmic reticulum lumen. They are expressed in nearly all cells. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon 1 encodes the leader peptide, exon 2 and 3 encode the alpha1 and alpha2 domains, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region and exons 6 and 7 encode the cytoplasmic tail. Polymorphisms within exon 2 and exon 3 are responsible for the peptide binding specificity of each class one molecule. Typing for these polymorphisms is routinely done for bone marrow and kidney transplantation. Hundreds of HLA-B alleles have been described. [provided by RefSeq, Jul 2008] |
Target gene [HLA-B] related to virus gene/protein/region 
Mutation Table: if previous studies reported that target gene was related to virus gene/region/protein, the information in this literature was listed in this section
| ID |
PMID |
Mutation |
Gene/Protein/Region |
Encoding Gene/Protein |
Disease |
Description |
Detail |
| 1 |
38397027 |
H47Y |
ORF7a
|
ORF7a
|
|
G
R
S
T
|
View |
Target gene [HLA-B] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE167075
|
MeRIP-seq;RNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens; Severe acute respiratory syndrome coronavirus 2);Illumina NovaSeq 6000 (Chlorocebus aethiops; Severe acute respiratory syndrome coronavirus 2) |
|
S GSE164485
|
scRNA-seq |
36 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE190680
|
RNA-seq |
100 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE152641
|
RNA-seq |
86 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE283771
|
RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE211934
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE163959
|
RNA-seq |
48 |
Illumina NextSeq 500 (Homo sapiens) |
|
S GSE206147
|
scRNA-seq |
1 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE240694
|
scRNA-seq |
9 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE166253
|
RNA-seq |
26 |
HiSeq X Ten (Homo sapiens) |
|
GSE153218
|
RNA-seq |
16 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE162323
|
RNA-seq |
42 |
Illumina NovaSeq 6000 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens; Severe acute respiratory syndrome coronavirus 2) |
|
GSE199816
|
RNA-seq |
27 |
DNBSEQ-G400 (Homo sapiens) |
|
GSE215262
|
RNA-seq |
25 |
Illumina NovaSeq 6000 (Homo sapiens) |
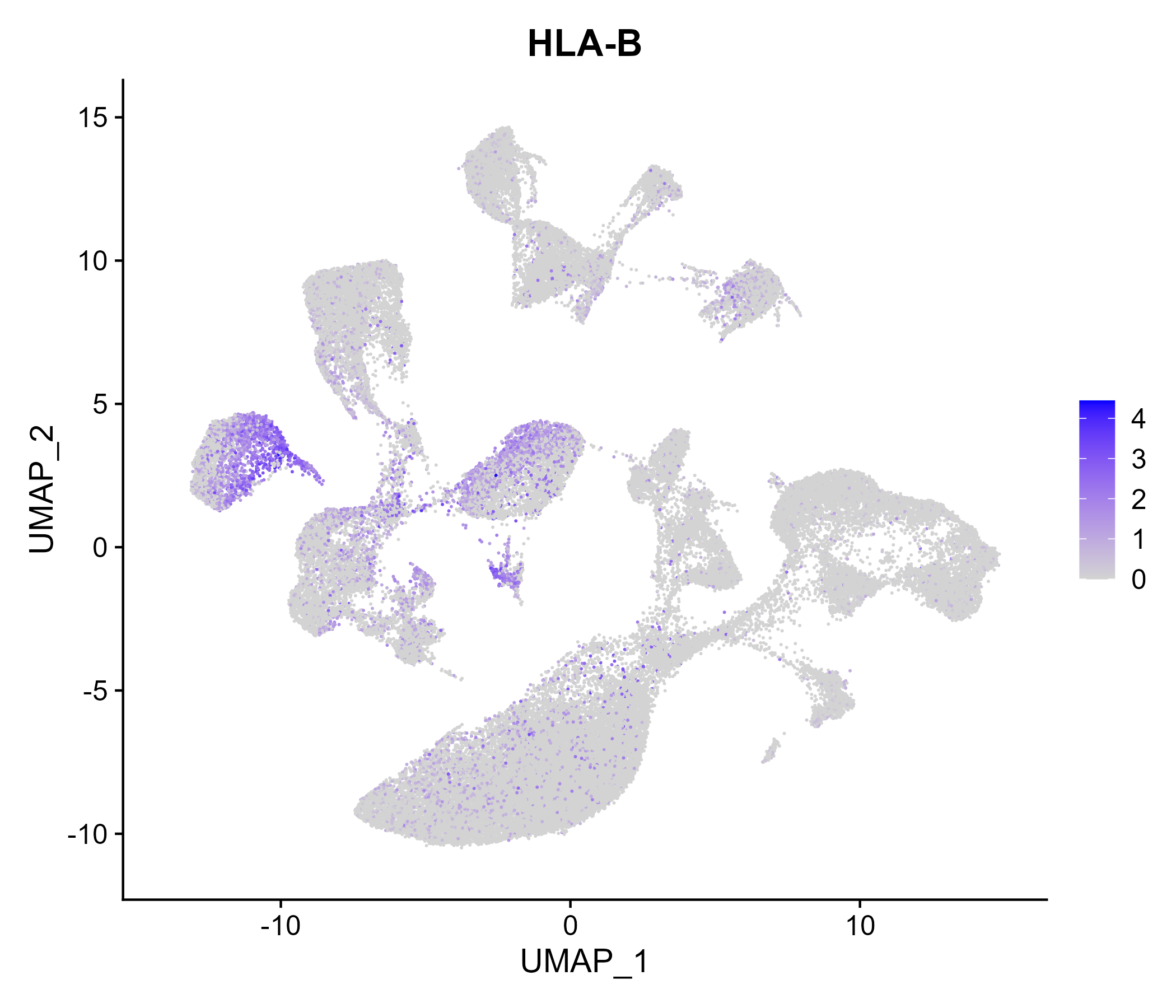
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE164485 - Gene expression in cell subsets
|
|
|
|
|
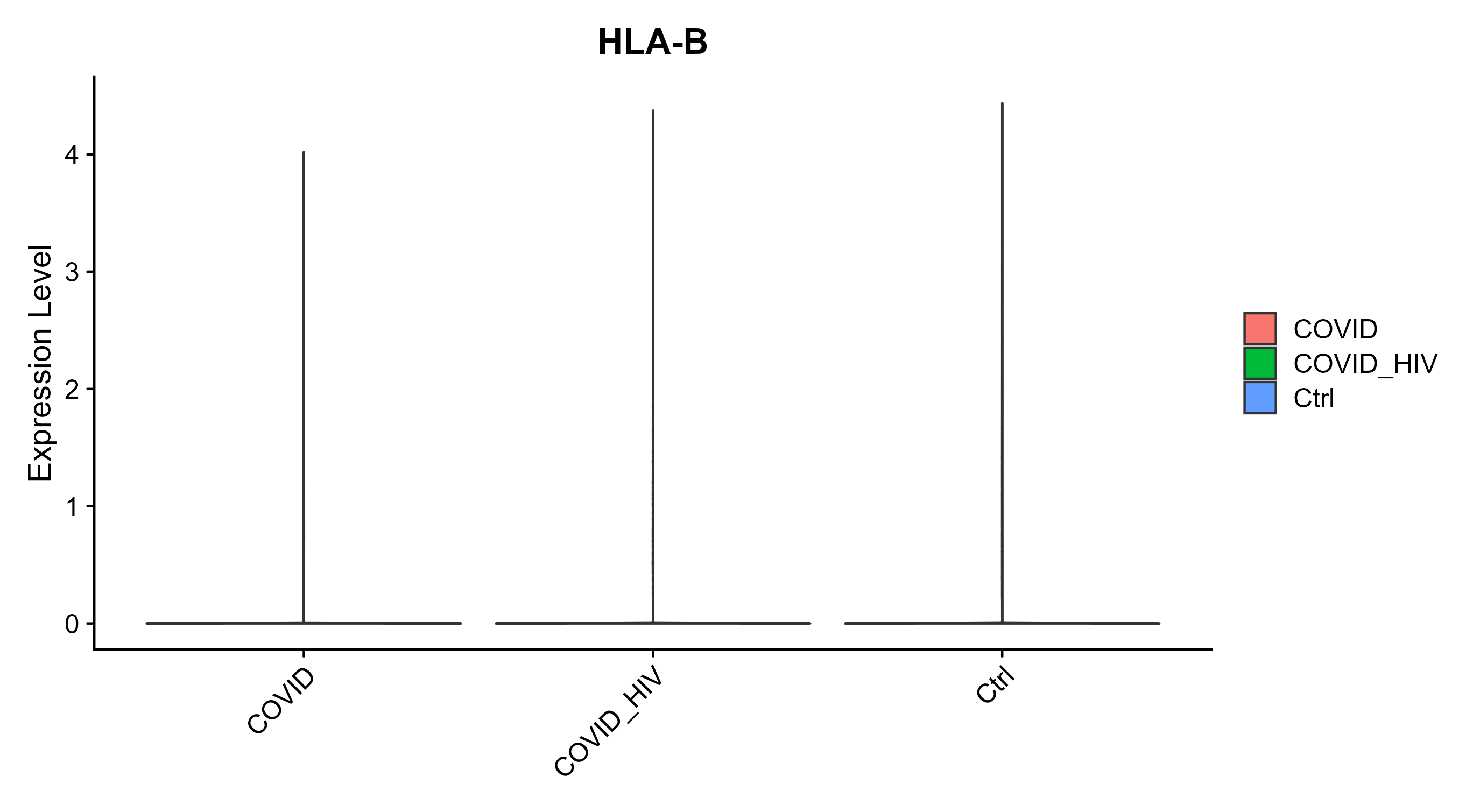
|
> Dataset: GSE206147 - Gene expression in cell subsets
|
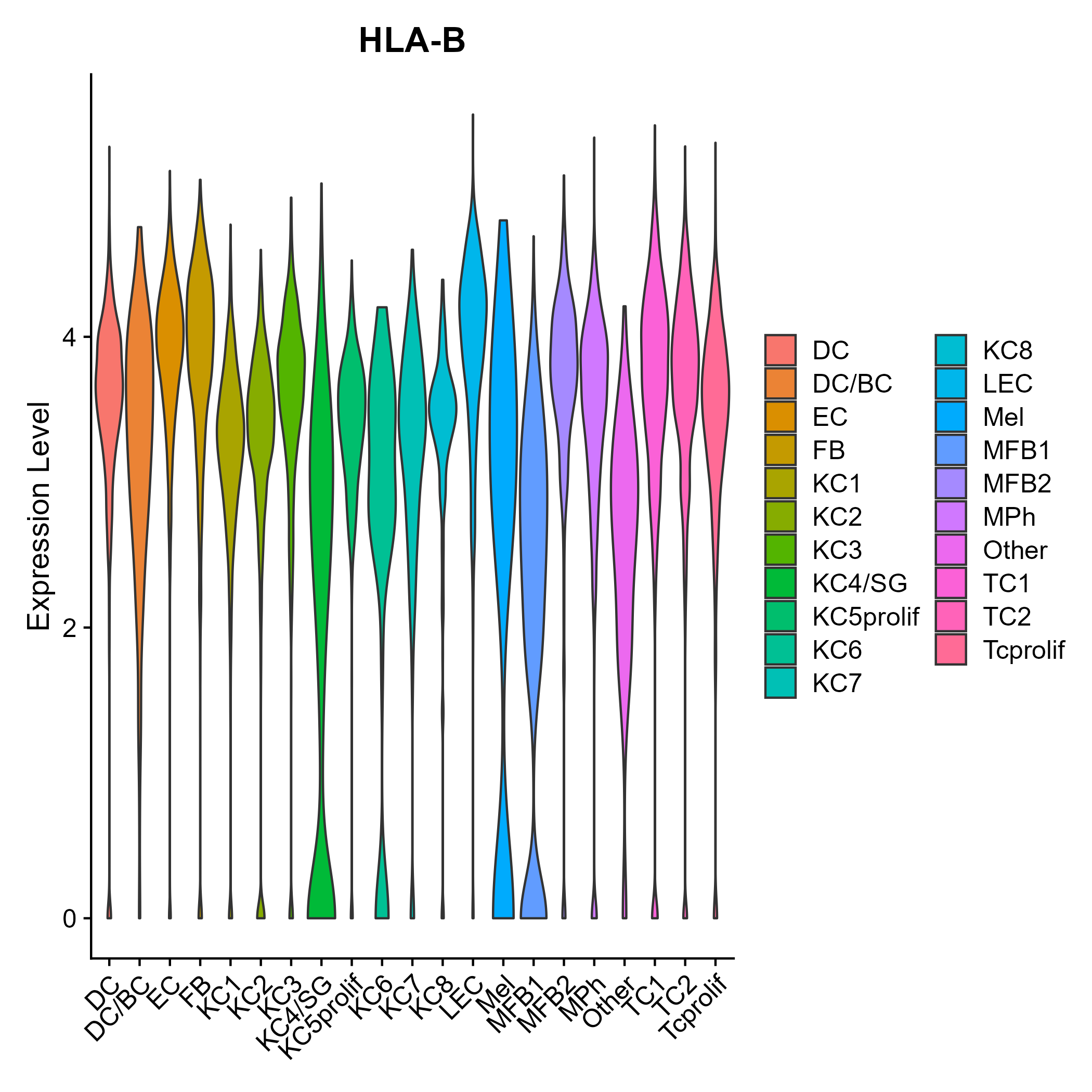
|
|
|
 SARS-CoV-2 Target gene Detail Information
SARS-CoV-2 Target gene Detail Information