Gene Information
|
Gene Name
|
INSR |
|
Gene ID
|
3643
|
|
Gene Full Name
|
insulin receptor |
|
Gene Alias
|
CD220|HHF5 |
|
Transcripts
|
ENSG00000171105
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles, Plasma membrane;Primary cilium transition zone, Centriolar satellite, Basal body; 
|
|
Membrane Info
|
CD markers, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins, RAS pathway related proteins |
|
Uniport_ID
|
P06213
|
|
HGNC ID
|
HGNC:6091
|
|
OMIM ID
|
147670 |
|
Summary
|
This gene encodes a member of the receptor tyrosine kinase family of proteins. The encoded preproprotein is proteolytically processed to generate alpha and beta subunits that form a heterotetrameric receptor. Binding of insulin or other ligands to this receptor activates the insulin signaling pathway, which regulates glucose uptake and release, as well as the synthesis and storage of carbohydrates, lipids and protein. Mutations in this gene underlie the inherited severe insulin resistance syndromes including type A insulin resistance syndrome, Donohue syndrome and Rabson-Mendenhall syndrome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2015] |
Target gene [INSR] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5003379 |
chr19 |
3 |
1 |
0 |
4 |
View |
| 5005907 |
chr19 |
55 |
4 |
1 |
60 |
View |
| 5006818 |
chr19 |
6 |
2 |
8 |
16 |
View |
| 5006819 |
chr19 |
18 |
4 |
3 |
25 |
View |
| 5015345 |
chr19 |
0 |
0 |
0 |
0 |
View |
Target gene [INSR] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE139324
|
scRNA-seq |
63 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
S GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
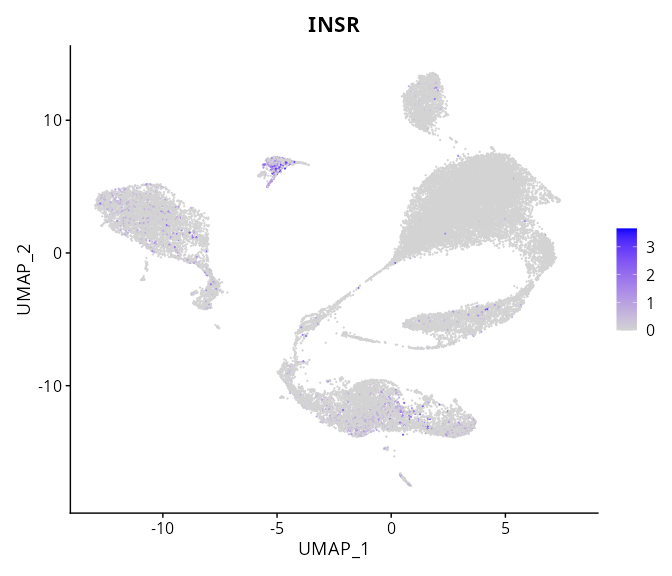
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE226620 - Gene expression in cell subsets
|
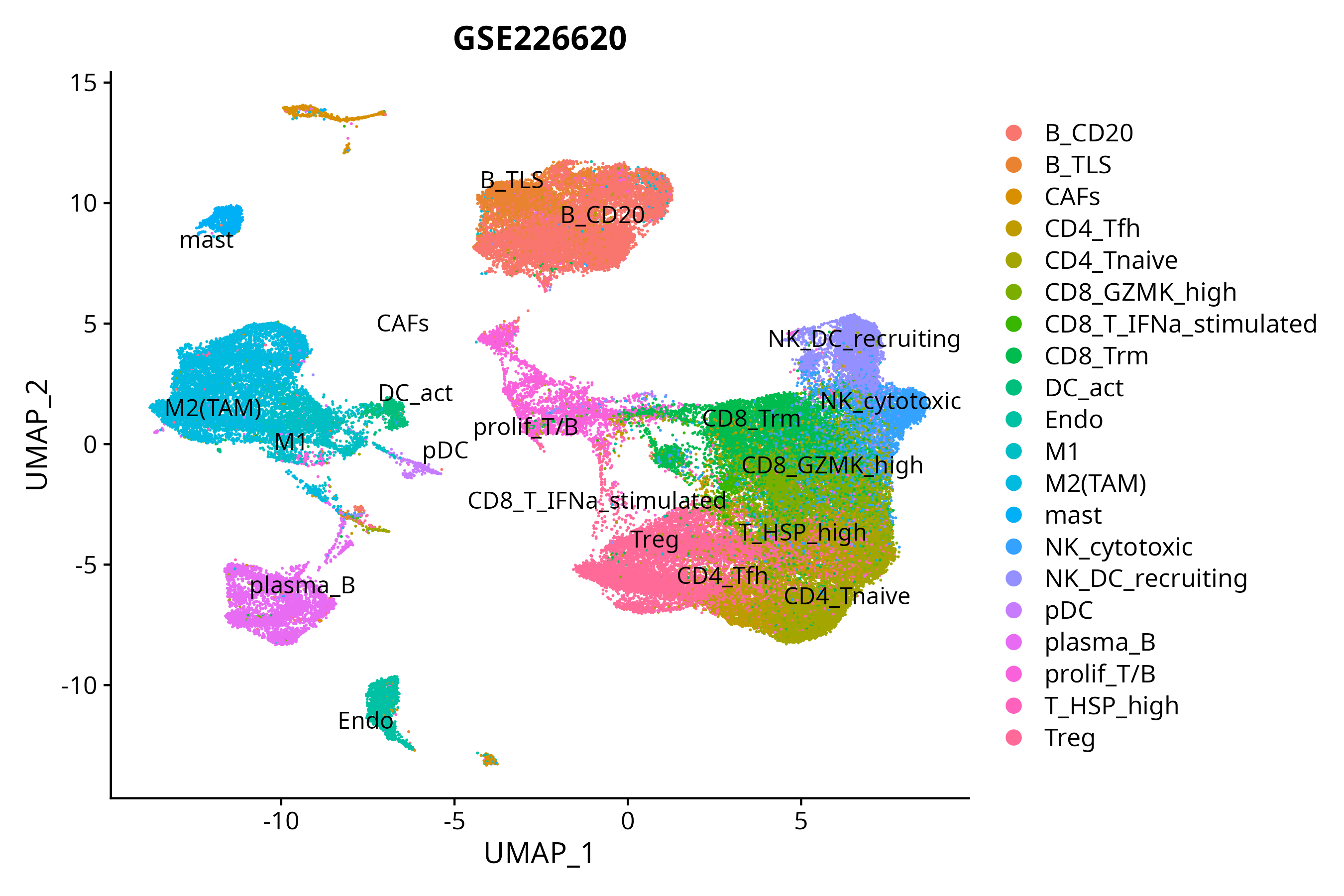
|
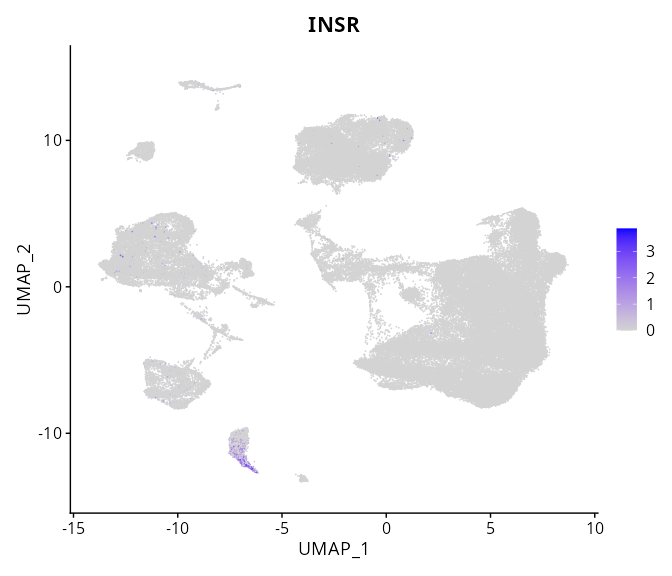
|

|
|
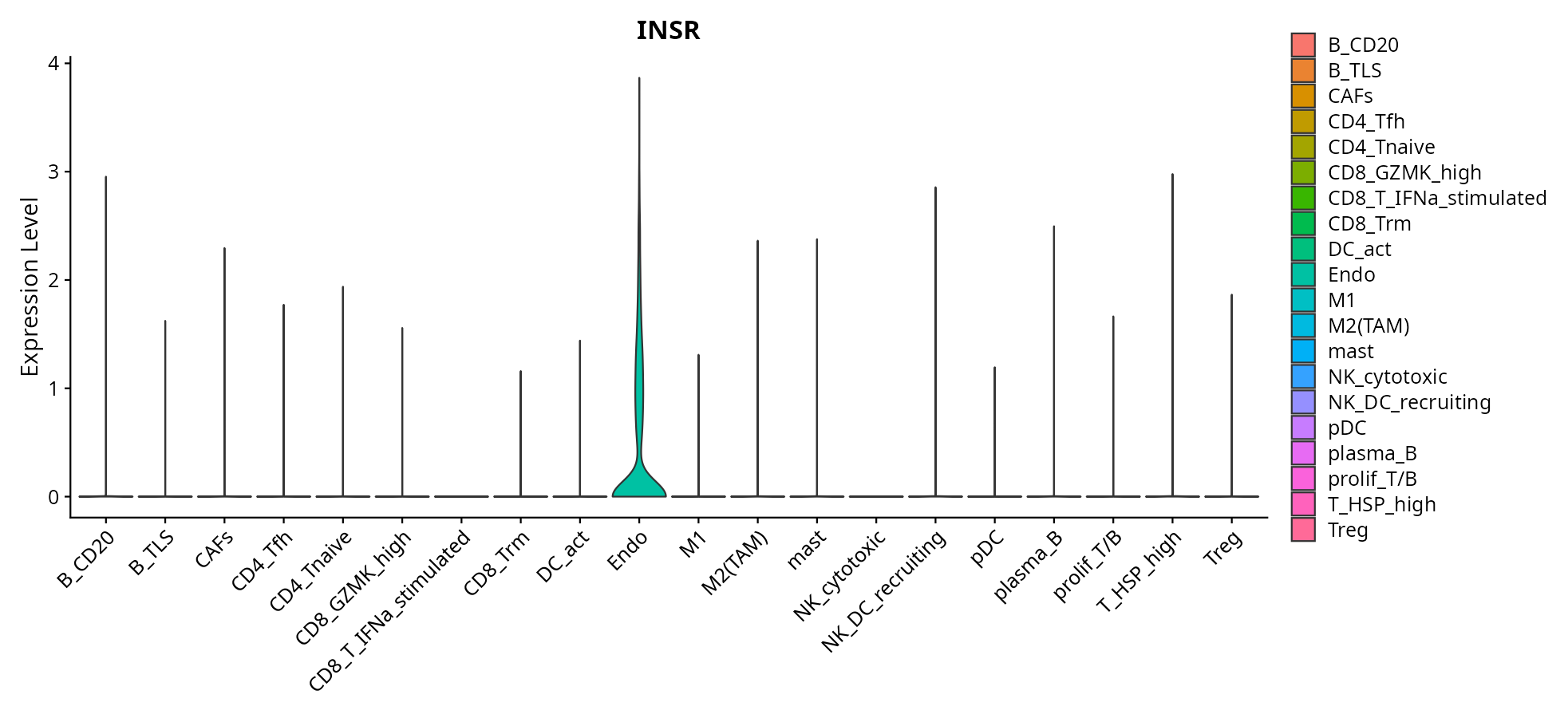
|
> Dataset: GSE164690 - Gene expression in cell subsets
|
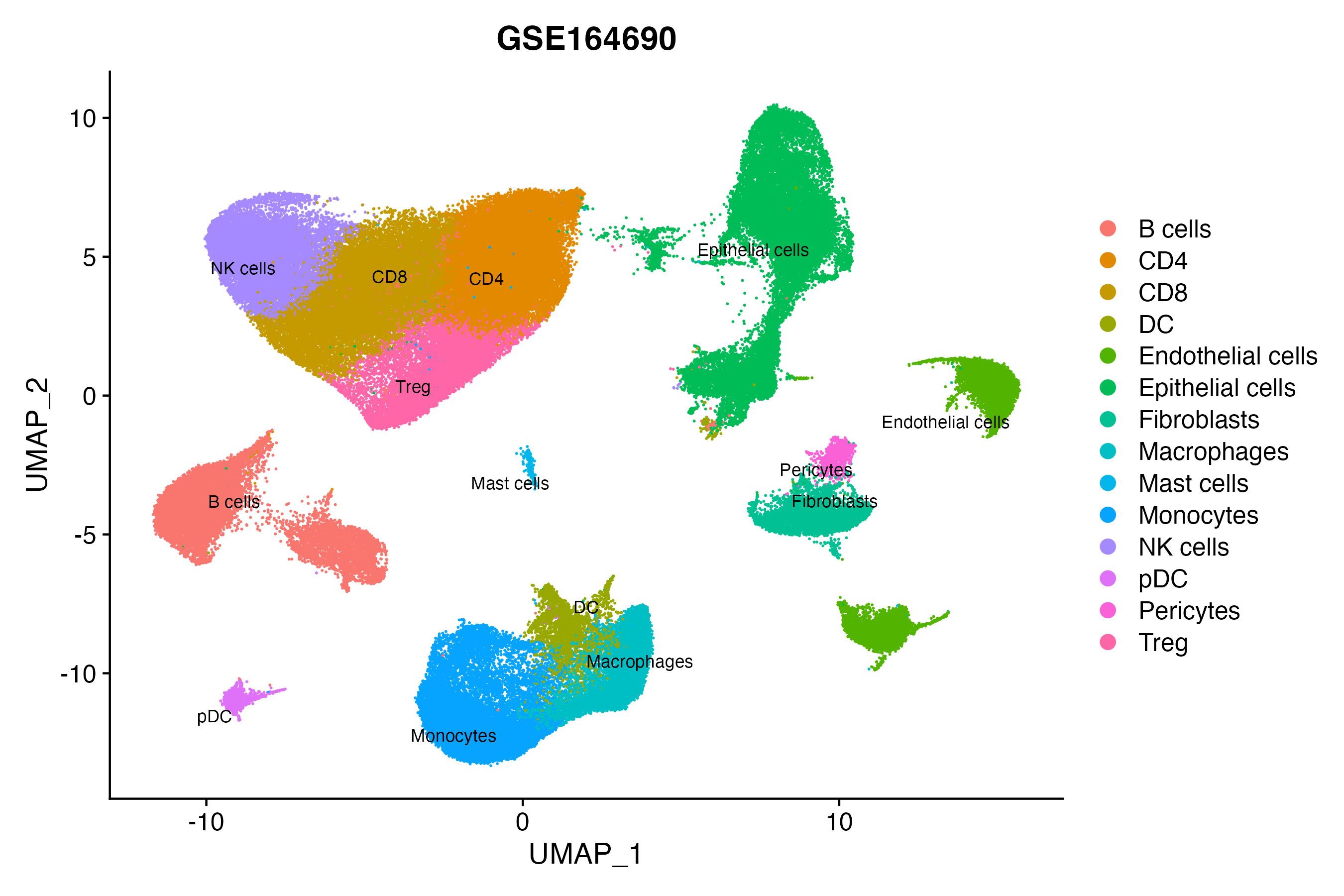
|
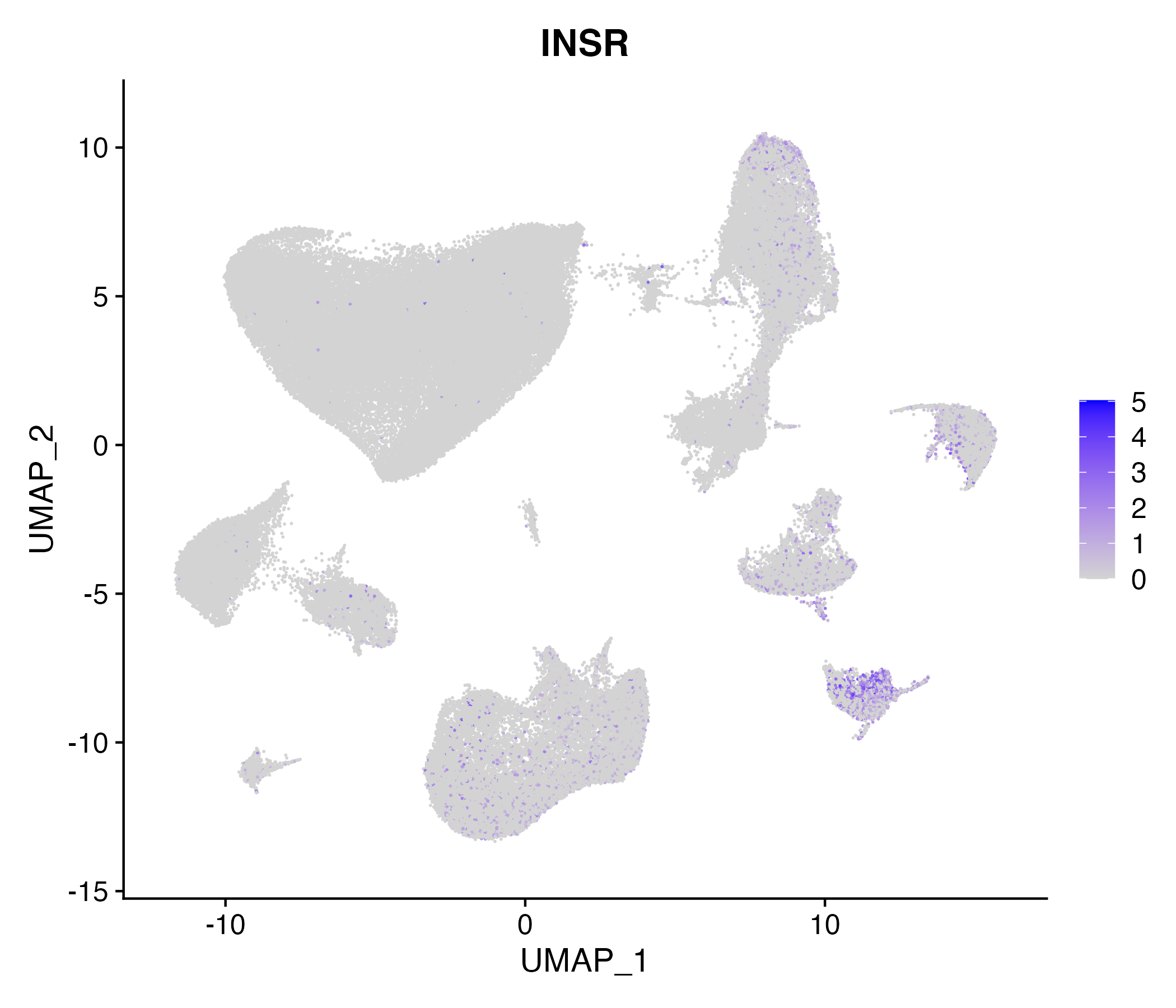
|

|
|
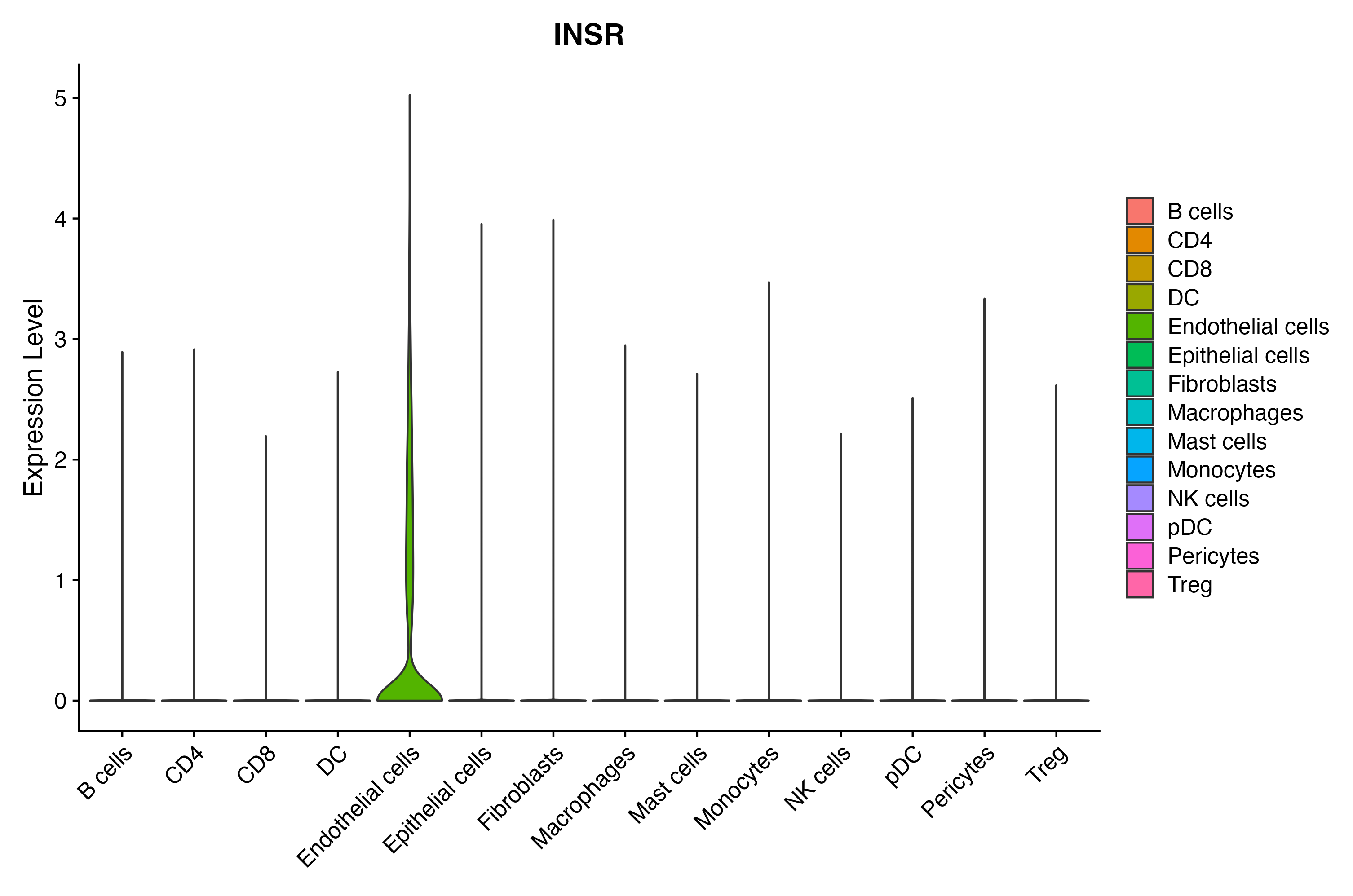
|
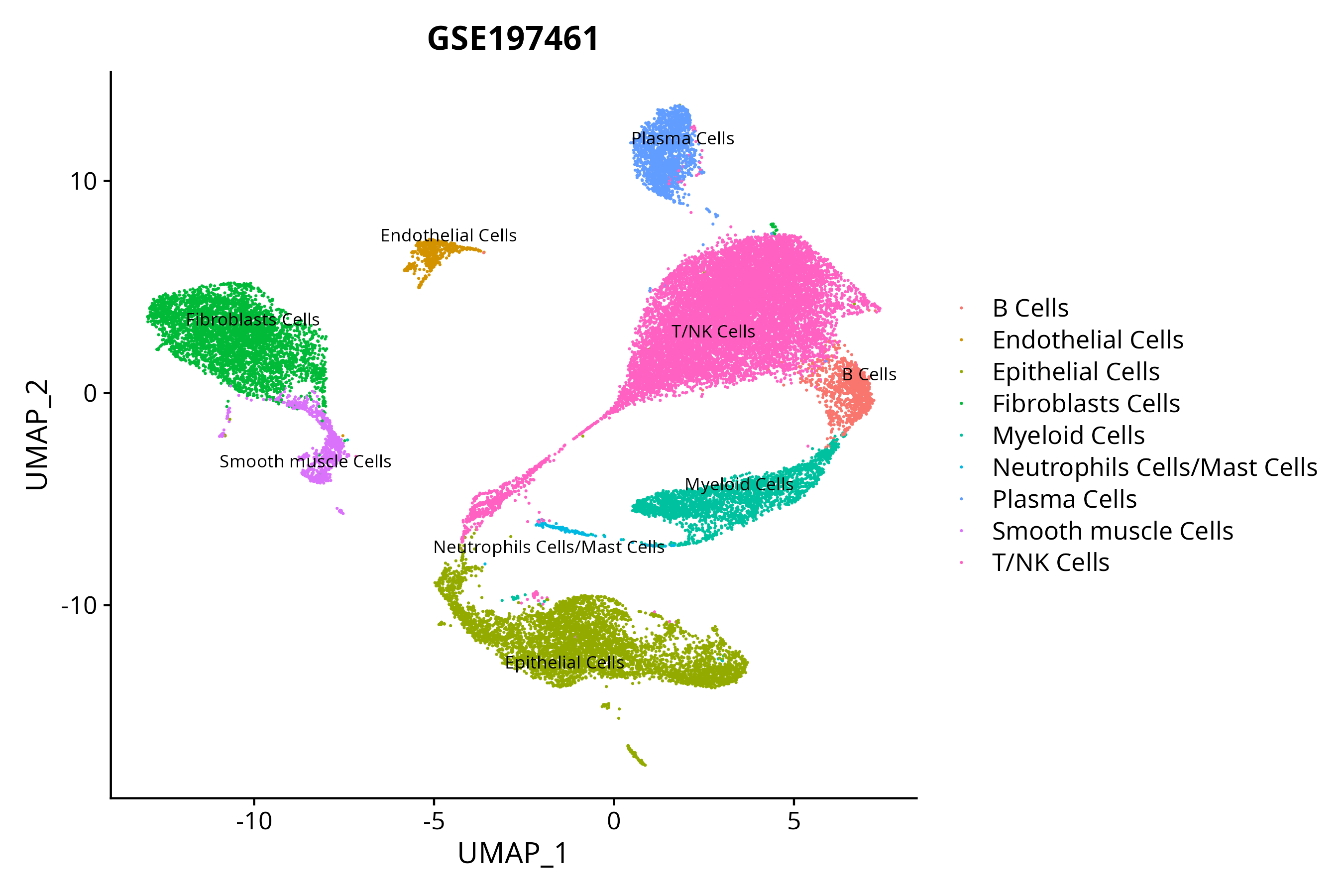
> Dataset: GSE197461 - Gene expression in cell subsets
|
|
|
|
|
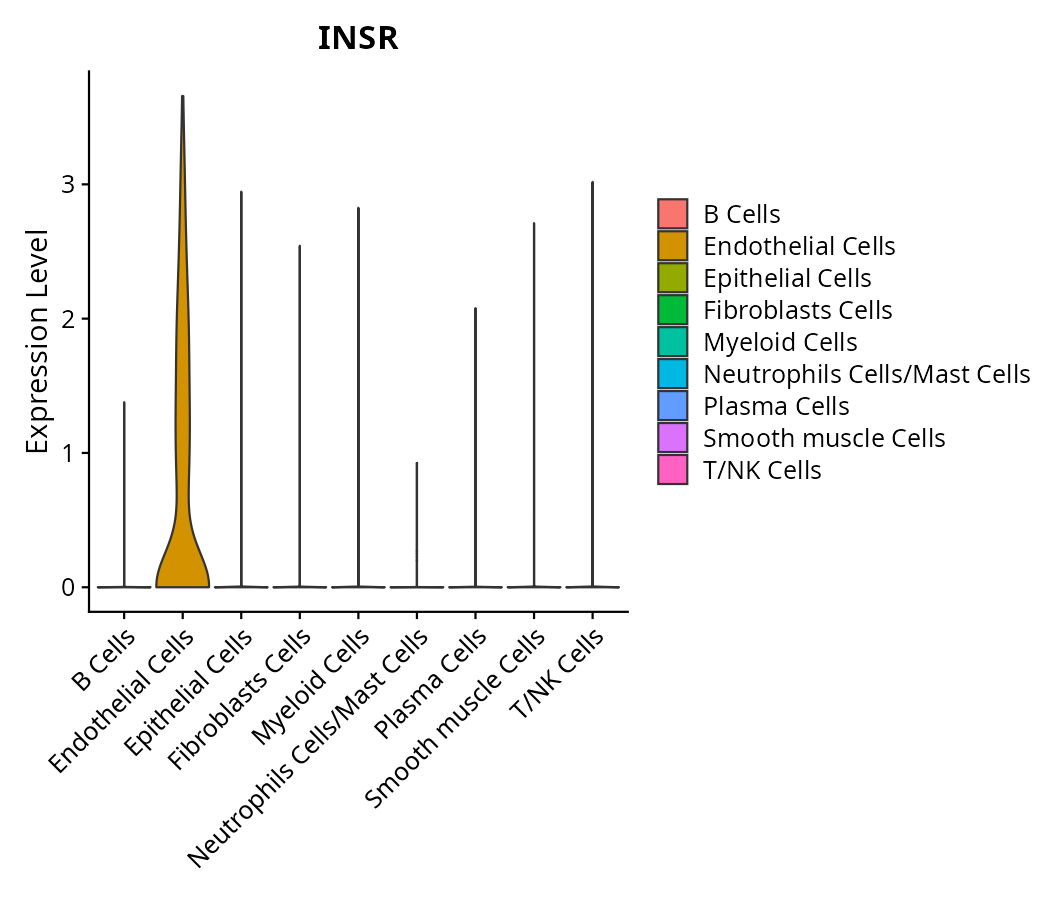
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information