Gene Information
|
Gene Name
|
KDR |
|
Gene ID
|
3791
|
|
Gene Full Name
|
kinase insert domain receptor |
|
Gene Alias
|
CD309|FLK1|VEGFR|VEGFR2 |
|
Transcripts
|
ENSG00000128052
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles;Nuclear membrane, Cytosol, Connecting piece, Mid piece, Principal piece;Intracellular and membrane; 
|
|
Membrane Info
|
Cancer-related genes, CD markers, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted membrane proteins, RAS pathway related proteins |
|
Uniport_ID
|
P35968
|
|
HGNC ID
|
HGNC:6307
|
|
OMIM ID
|
191306 |
|
Summary
|
Vascular endothelial growth factor (VEGF) is a major growth factor for endothelial cells. This gene encodes one of the two receptors of the VEGF. This receptor, known as kinase insert domain receptor, is a type III receptor tyrosine kinase. It functions as the main mediator of VEGF-induced endothelial proliferation, survival, migration, tubular morphogenesis and sprouting. The signalling and trafficking of this receptor are regulated by multiple factors, including Rab GTPase, P2Y purine nucleotide receptor, integrin alphaVbeta3, T-cell protein tyrosine phosphatase, etc.. Mutations of this gene are implicated in infantile capillary hemangiomas. [provided by RefSeq, May 2009] |
Target gene [KDR] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5006721 |
chr4 |
2 |
0 |
0 |
2 |
View |
Target gene [KDR] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
S GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
M TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE199029
|
Variation;RNA-seq |
44 |
NanoString Human nCounter PanCancer IO360 Panel |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
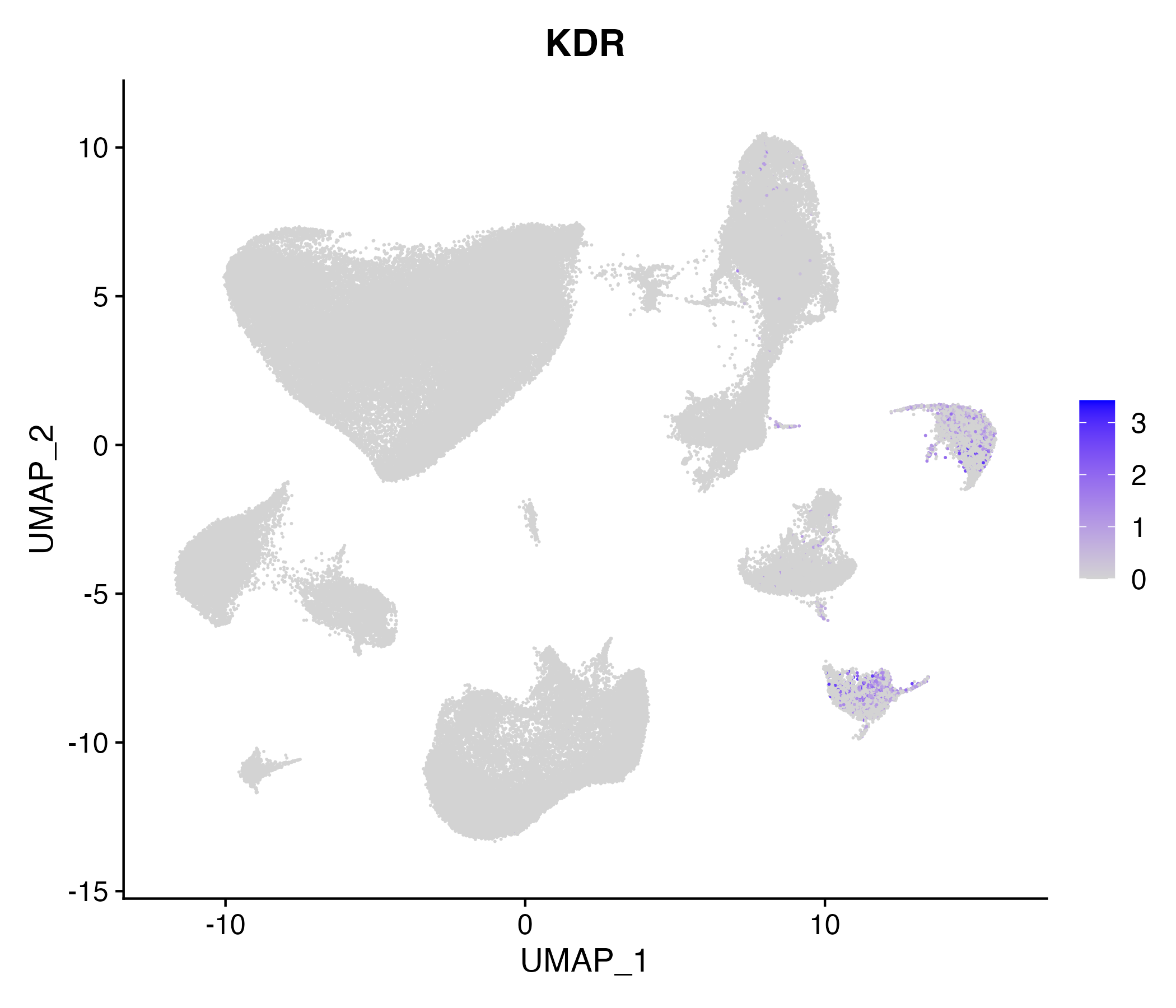
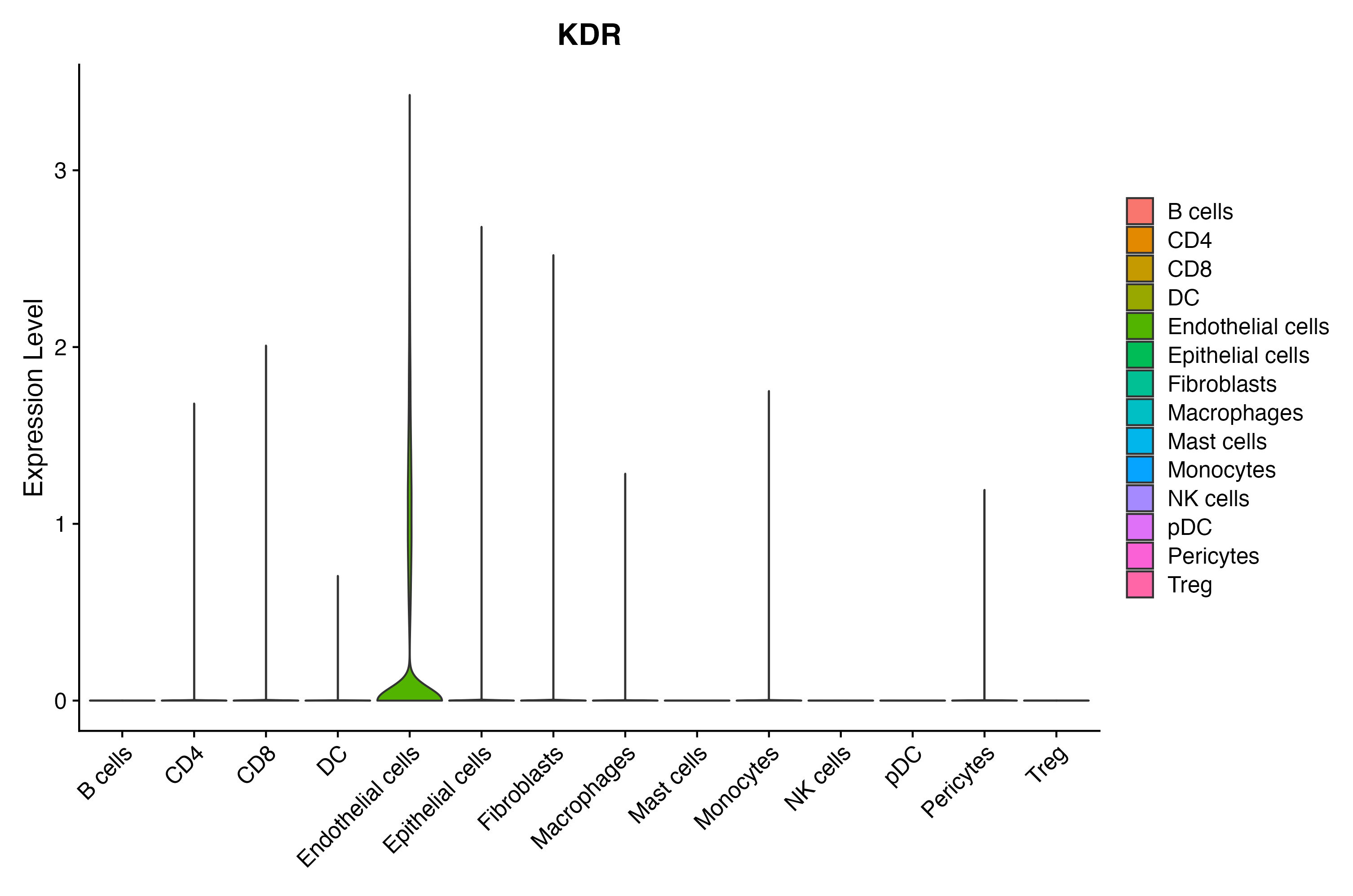
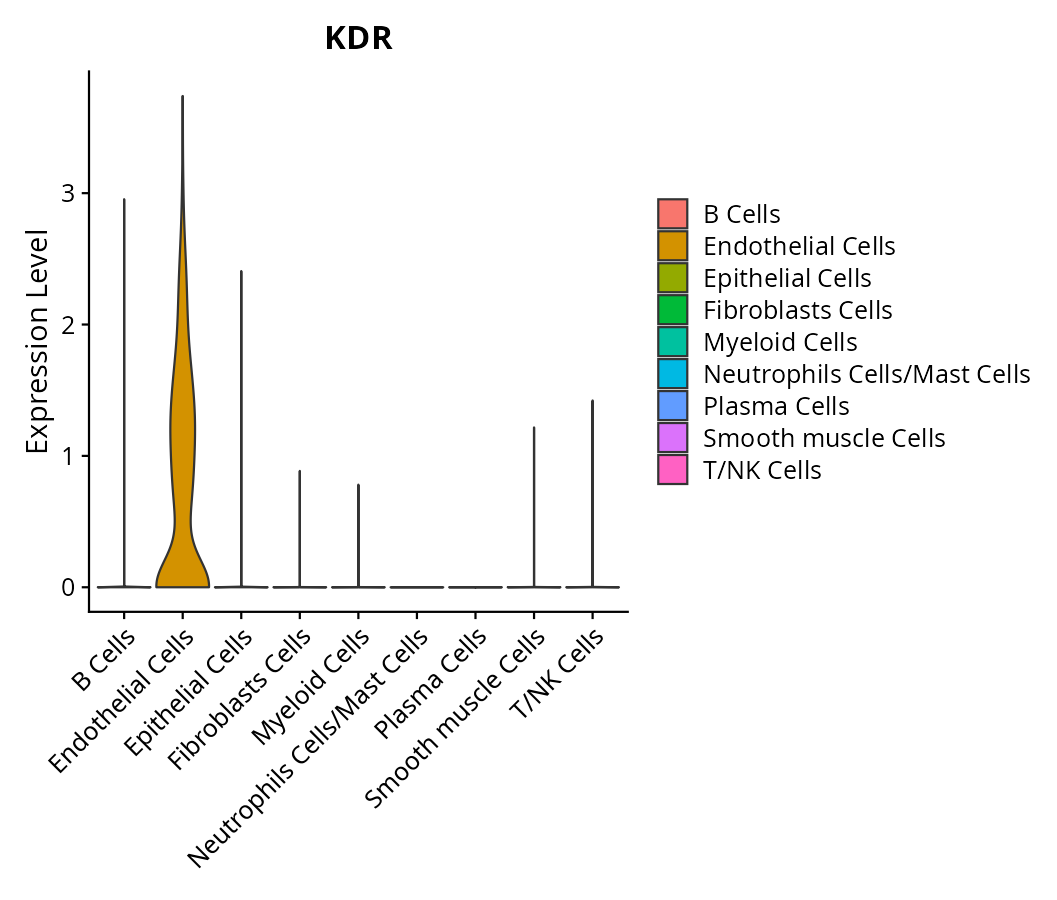
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE164690 - Gene expression in cell subsets
|
|
|

|
|
|
> Dataset: GSE197461 - Gene expression in cell subsets
|
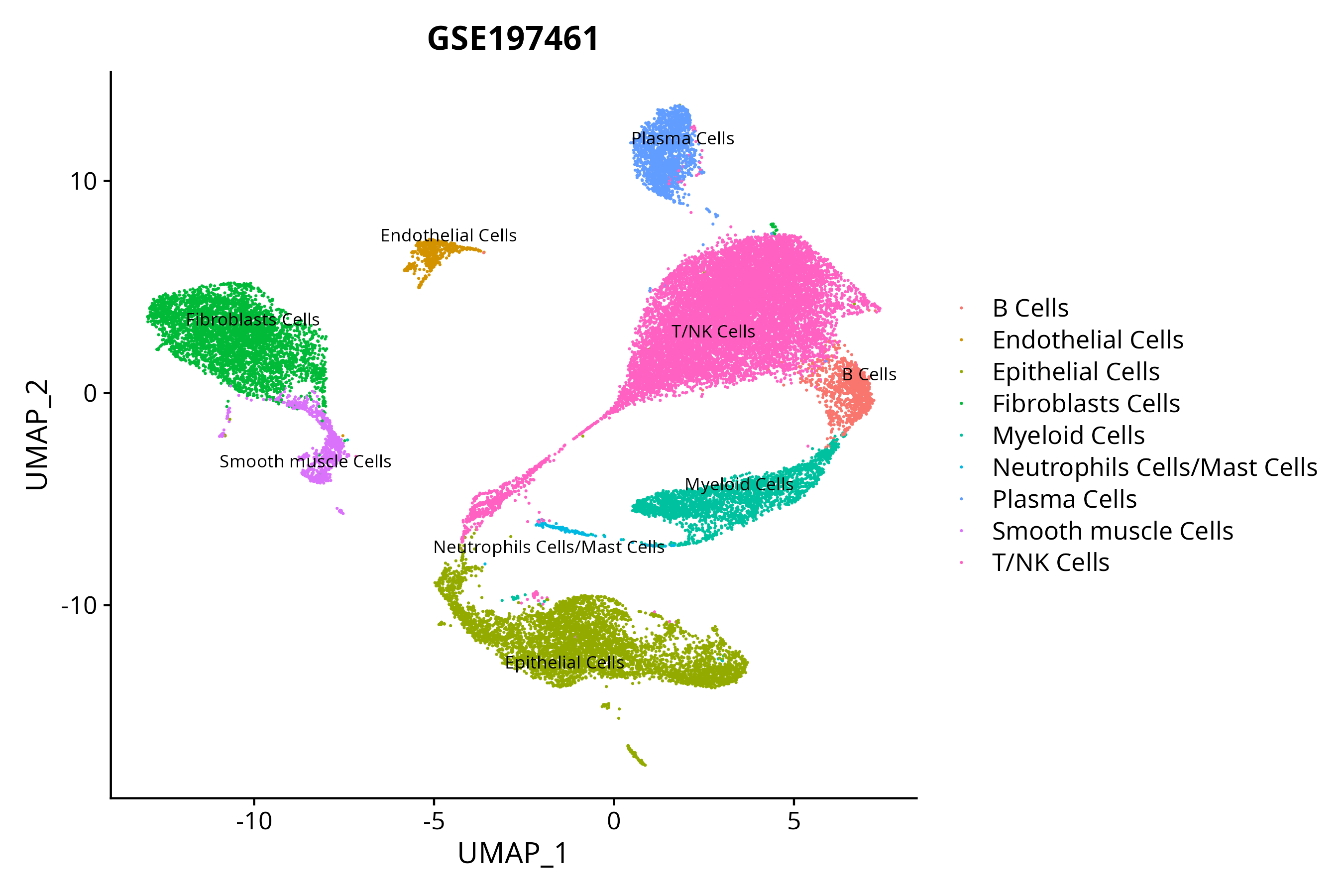
|
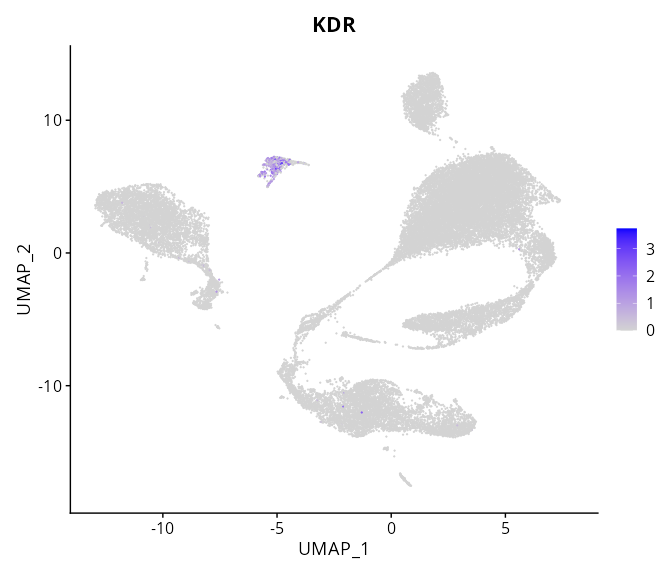
|
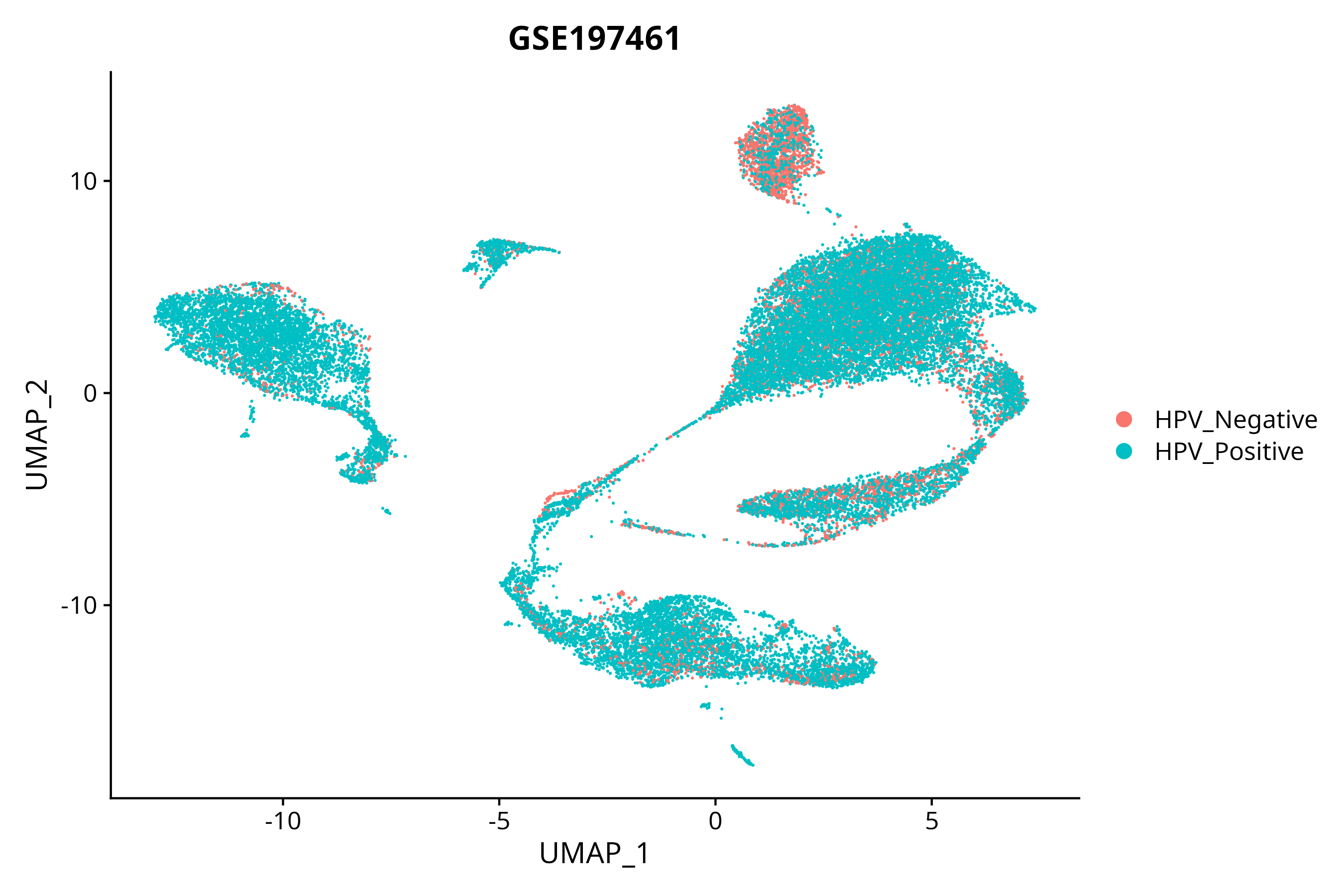
|
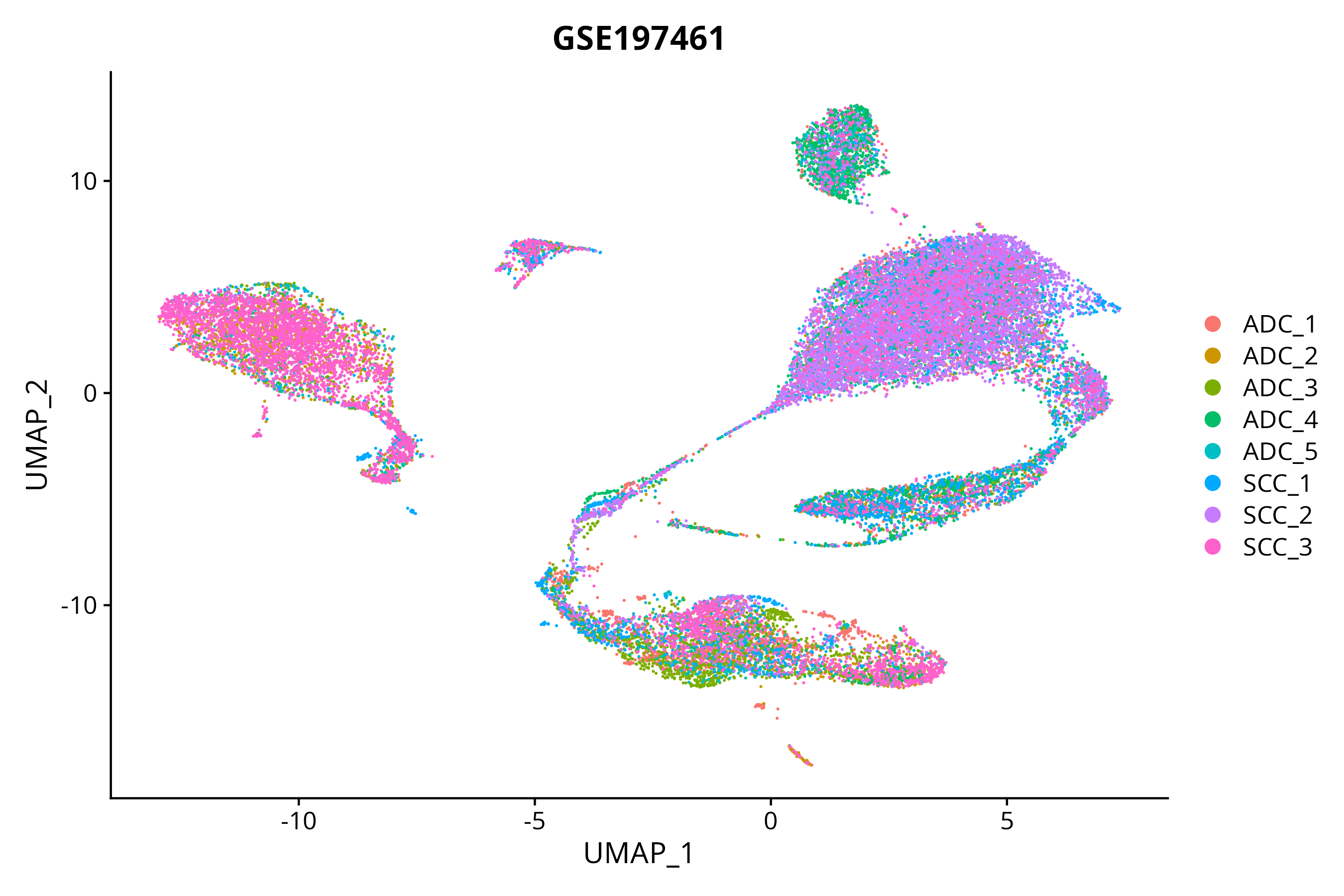
|
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information