Gene Information
|
Gene Name
|
TIAM1 |
|
Gene ID
|
7074
|
|
Gene Full Name
|
TIAM Rac1 associated GEF 1 |
|
Gene Alias
|
NEDLDS|TIAM-1 |
|
Transcripts
|
ENSG00000156299
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm, Cell Junctions;Nuclear membrane, Cytokinetic bridge, Cytosol; 
|
|
Membrane Info
|
Disease related genes, Plasma proteins, Predicted intracellular proteins, RAS pathway related proteins |
|
Uniport_ID
|
Q13009
|
|
HGNC ID
|
HGNC:11805
|
|
OMIM ID
|
600687 |
|
Summary
|
This gene encodes a RAC1-specific guanine nucleotide exchange factor (GEF). GEFs mediate the exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP). The binding of GTP induces a conformational change in RAC1 that allows downstream effectors to bind and transduce a signal. This gene thus regulates RAC1 signaling pathways that affect cell shape, migration, adhesion, growth, survival, and polarity, as well as influencing actin cytoskeletal formation, endocytosis, and membrane trafficking. This gene thus plays an important role in cell invasion, metastasis, and carcinogenesis. In addition to RAC1, the encoded protein activates additional Rho-like GTPases such as CDC42, RAC2, RAC3 and RHOA. This gene encodes multiple protein isoforms that experience a diverse array of intramolecular, protein-protein, and phosphorylation interactions as well as phosphoinositide binding. Both the longer and shorter isoforms have C-terminal Dbl homology (DH) and pleckstrin homology (PH) domains while only the longer isoforms of this gene have the N-terminal myristoylation site and the downstream N-terminal PH domain, ras-binding domain (RBD), and PSD-95/DlgA/ZO-1 (PDZ) domain. [provided by RefSeq, Jul 2017] |
Target gene [TIAM1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5006420 |
chr21 |
11 |
0 |
0 |
11 |
View |
| 5008080 |
chr21 |
4 |
2 |
0 |
6 |
View |
| 5010921 |
chr21 |
1 |
0 |
0 |
1 |
View |
| 5015709 |
chr21 |
11 |
2 |
1 |
14 |
View |
Target gene [TIAM1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: TCGA_CESC - TIAM1 expression across samples
|
Volcano Plot
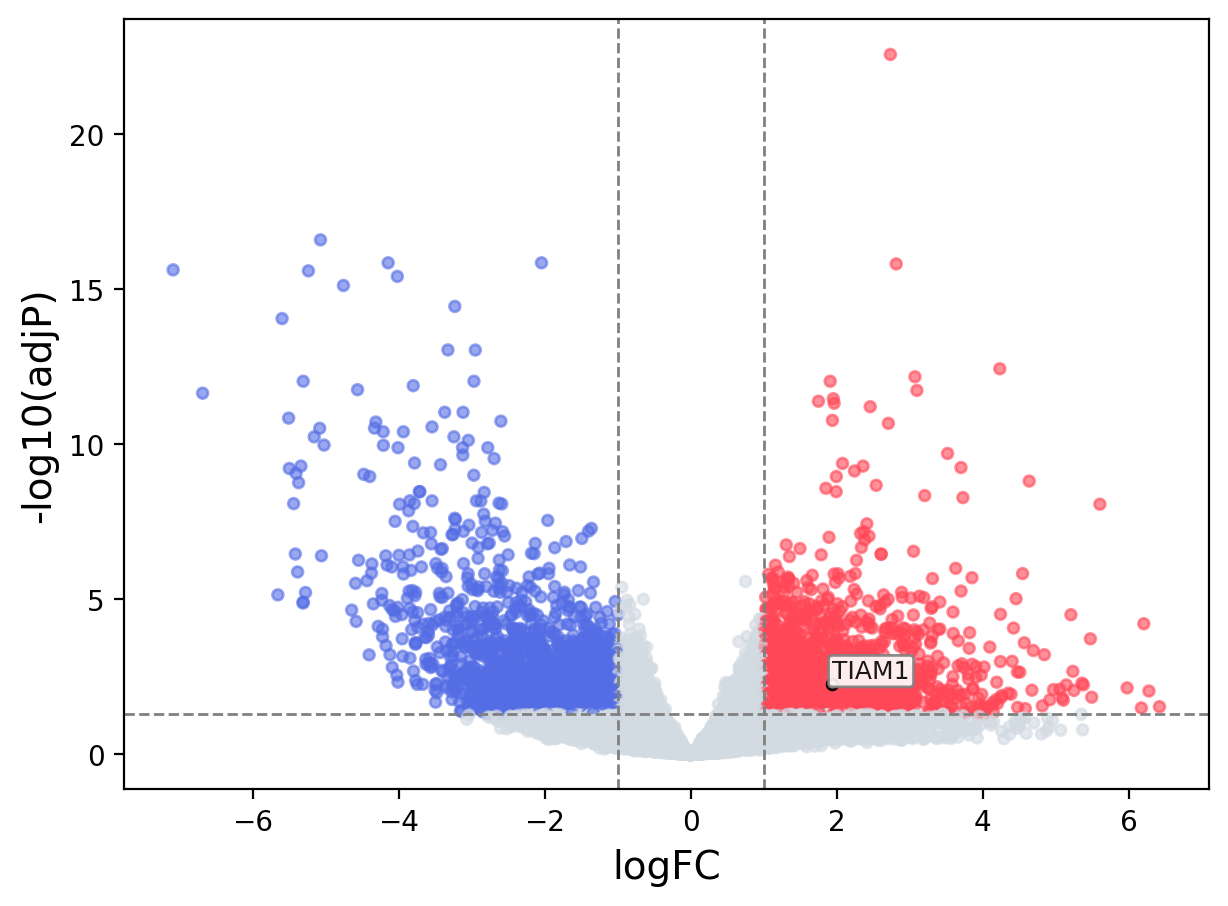
|
Bar Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information