Gene Information
|
Gene Name
|
CXCR6 |
|
Gene ID
|
10663
|
|
Gene Full Name
|
C-X-C motif chemokine receptor 6 |
|
Gene Alias
|
BONZO|CD186|CDw186|STRL33|TYMSTR |
|
Transcripts
|
ENSG00000172215
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
CD markers, G-protein coupled receptors, Predicted membrane proteins |
|
Uniport_ID
|
O00574
|
|
HGNC ID
|
HGNC:16647
|
|
OMIM ID
|
605163 |
|
Summary
|
The protein encoded by this gene is a G protein-coupled receptor with seven transmembrane domains that belongs to the CXC chemokine receptor family. This family also includes CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, and CXCR7. This gene, which maps to the chemokine receptor gene cluster, is expressed in several T lymphocyte subsets and bone marrow stromal cells. The encoded protein and its exclusive ligand, chemokine ligand 16 (CCL16), are part of a signalling pathway that regulates T lymphocyte migration to various peripheral tissues (the liver, spleen red pulp, intestine, lungs, and skin) and promotes cell-cell interaction with dendritic cells and fibroblastic reticular cells. CXCR6/CCL16 also controls the localization of resident memory T lymphocytes to different compartments of the lung and maintains airway resident memory T lymphocytes, which are an important first line of defense against respiratory pathogens. The encoded protein serves as an entry coreceptor used by HIV-1 and SIV to enter target cells, in conjunction with CD4. [provided by RefSeq, Aug 2020] |
Target gene [CXCR6] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5004776 |
chr3 |
62 |
8 |
27 |
97 |
View |
Target gene [CXCR6] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
S GSE139324
|
scRNA-seq |
63 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
S GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE199029
|
Variation;RNA-seq |
44 |
NanoString Human nCounter PanCancer IO360 Panel |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: TCGA_CESC - CXCR6 expression across samples
|
Volcano Plot
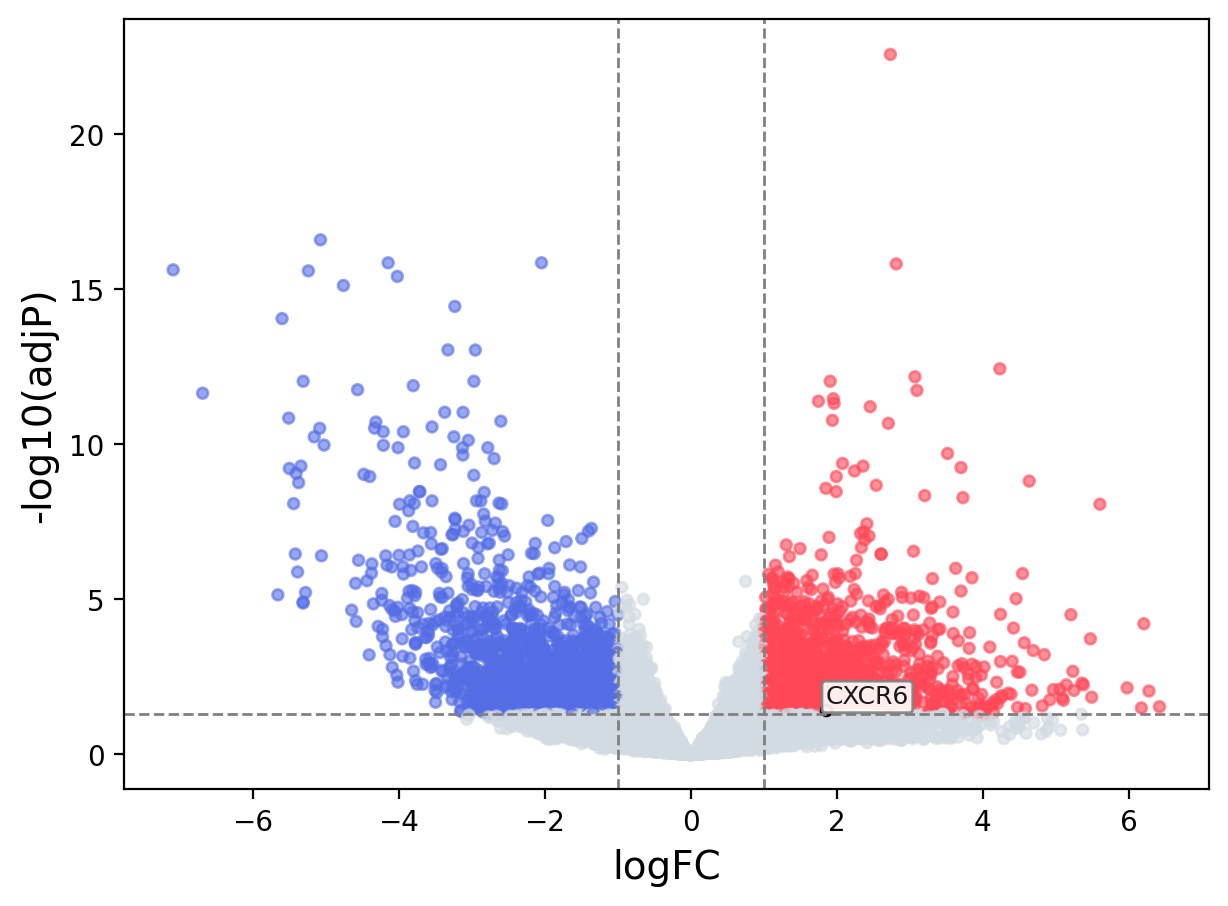
|
Bar Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information