Gene Information
|
Gene Name
|
ERBB2 |
|
Gene ID
|
2064
|
|
Gene Full Name
|
erb-b2 receptor tyrosine kinase 2 |
|
Gene Alias
|
CD340|HER-2|HER-2/neu|HER2|MLN 19|MLN-19|NEU|NGL|TKR1|VSCN2|c-ERB-2|c-ERB2|p185(erbB2) |
|
Transcripts
|
ENSG00000141736
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Nucleoplasm, Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, CD markers, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
P04626
|
|
HGNC ID
|
HGNC:3430
|
|
OMIM ID
|
164870 |
|
Summary
|
This gene encodes a member of the epidermal growth factor (EGF) receptor family of receptor tyrosine kinases. This protein has no ligand binding domain of its own and therefore cannot bind growth factors. However, it does bind tightly to other ligand-bound EGF receptor family members to form a heterodimer, stabilizing ligand binding and enhancing kinase-mediated activation of downstream signalling pathways, such as those involving mitogen-activated protein kinase and phosphatidylinositol-3 kinase. Allelic variations at amino acid positions 654 and 655 of isoform a (positions 624 and 625 of isoform b) have been reported, with the most common allele, Ile654/Ile655, shown here. Amplification and/or overexpression of this gene has been reported in numerous cancers, including breast and ovarian tumors. Alternative splicing results in several additional transcript variants, some encoding different isoforms and others that have not been fully characterized. [provided by RefSeq, Jul 2008] |
Target gene [ERBB2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
Target gene [ERBB2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE199029
|
Variation;RNA-seq |
44 |
NanoString Human nCounter PanCancer IO360 Panel |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
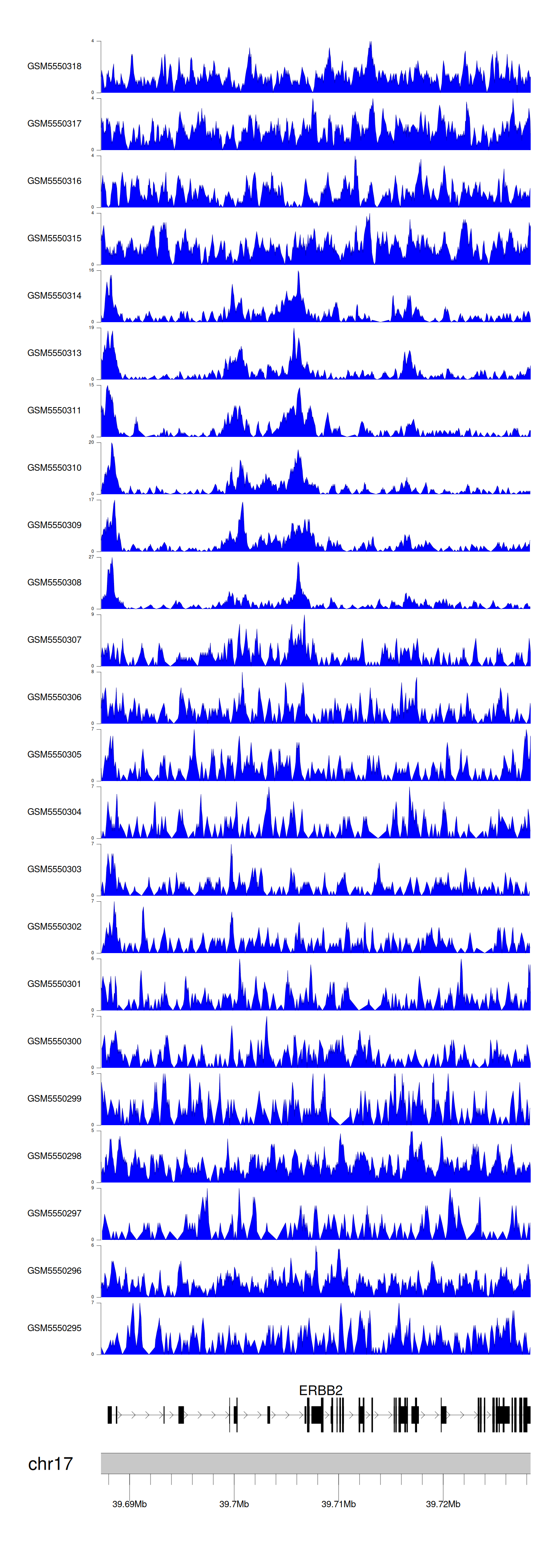
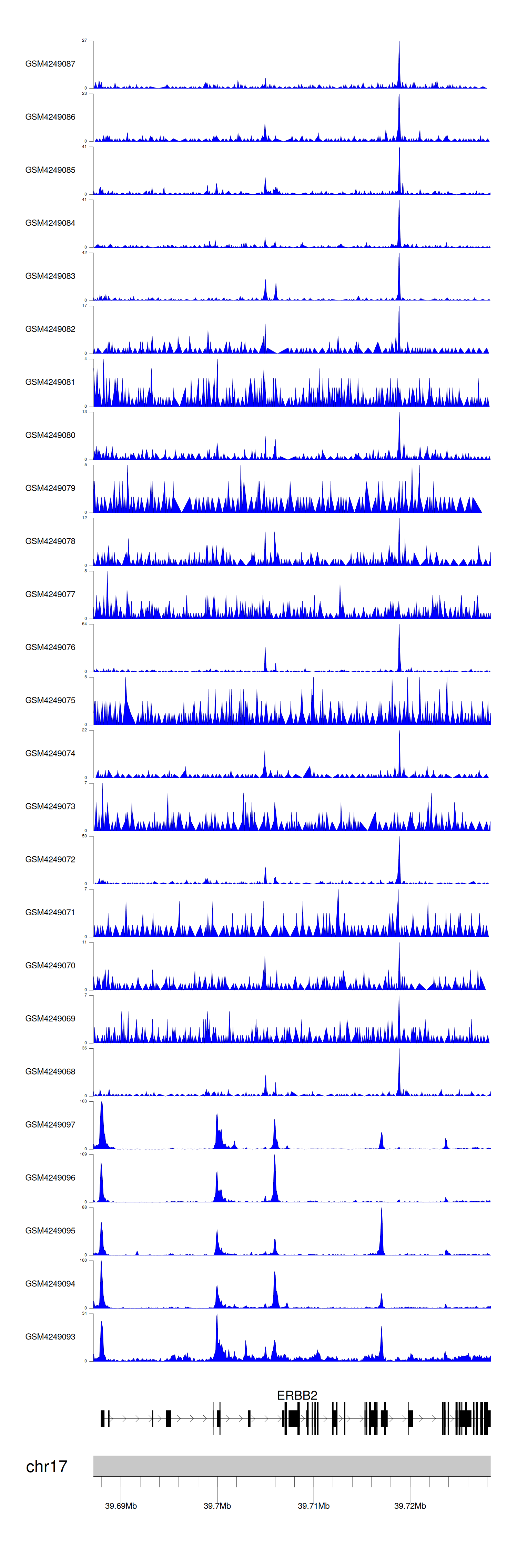
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE183048 - ERBB2 peak across samples
|
Peak Plot
|
> Dataset: GSE143026 - ERBB2 peak across samples
|
Peak Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information