Gene Information
|
Gene Name
|
FGF13 |
|
Gene ID
|
2258
|
|
Gene Full Name
|
fibroblast growth factor 13 |
|
Gene Alias
|
DEE90|FGF-13|FGF2|FHF-2|FHF2|LINC00889|XLID110 |
|
Transcripts
|
ENSG00000129682
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Cytosol; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Predicted intracellular proteins |
|
Uniport_ID
|
Q92913
|
|
HGNC ID
|
HGNC:3670
|
|
OMIM ID
|
300070 |
|
Summary
|
The protein encoded by this gene is a member of the fibroblast growth factor (FGF) family. FGF family members possess broad mitogenic and cell survival activities, and are involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth, and invasion. This gene is located in a region on chromosome X, which is associated with Borjeson-Forssman-Lehmann syndrome (BFLS), making it a possible candidate gene for familial cases of the BFLS, and for other syndromal and nonspecific forms of X-linked cognitive disability mapping to this region. Alternative splicing of this gene at the 5' end results in several transcript variants encoding different isoforms with different N-termini. [provided by RefSeq, Nov 2008] |
Target gene [FGF13] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5001569 |
chrX |
24 |
1 |
3 |
28 |
View |
| 5001614 |
chrX |
3 |
8 |
16 |
27 |
View |
| 5004479 |
chrX |
0 |
0 |
0 |
0 |
View |
| 5005274 |
chrX |
1 |
2 |
0 |
3 |
View |
| 5006056 |
chrX |
2 |
1 |
0 |
3 |
View |
| 5009091 |
chrX |
0 |
0 |
0 |
0 |
View |
| 5010659 |
chrX |
4 |
0 |
0 |
4 |
View |
| 5011431 |
chrX |
0 |
0 |
0 |
0 |
View |
Target gene [FGF13] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
E GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE199029
|
Variation;RNA-seq |
44 |
NanoString Human nCounter PanCancer IO360 Panel |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE55550 - FGF13 expression across samples
|
Volcano Plot
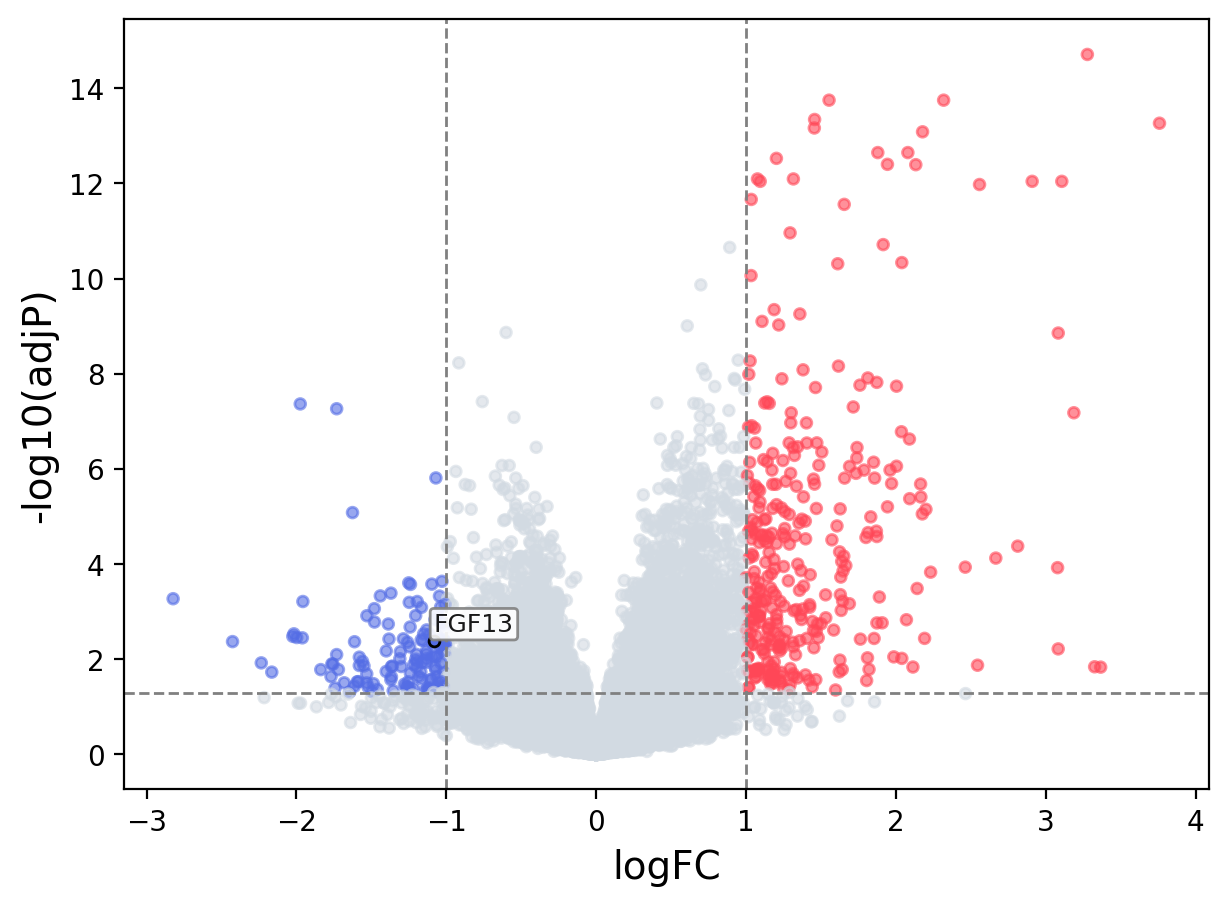
|
Bar Plot
|
> Dataset: TCGA_CESC - FGF13 expression across samples
|
Volcano Plot
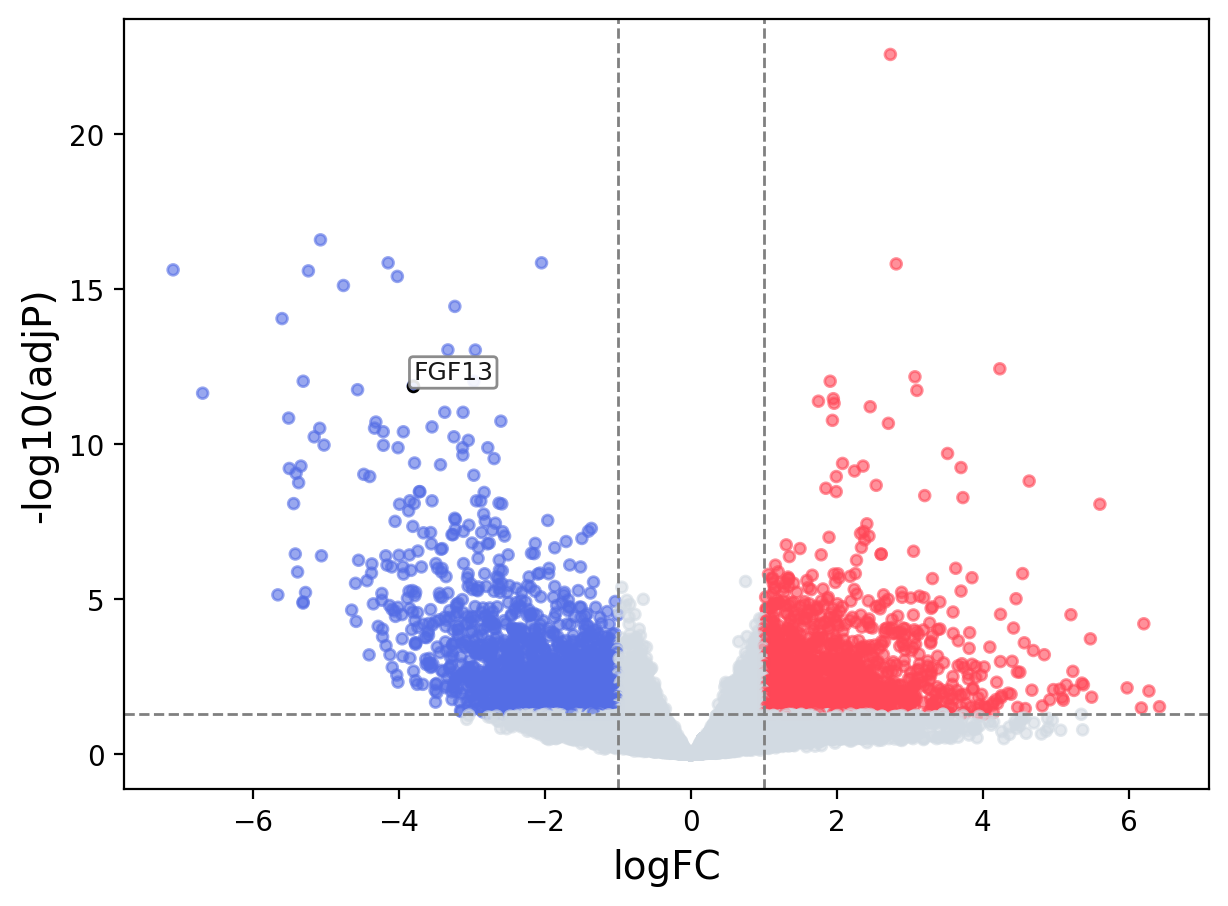
|
Bar Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information