Gene Information
|
Gene Name
|
MET |
|
Gene ID
|
4233
|
|
Gene Full Name
|
MET proto-oncogene, receptor tyrosine kinase |
|
Gene Alias
|
AUTS9|DA11|DFNB97|HGFR|RCCP2|c-Met |
|
Transcripts
|
ENSG00000105976
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Cytosol;Secreted to blood; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins, Predicted secreted proteins, RAS pathway related proteins, Transporters |
|
Uniport_ID
|
P08581
|
|
HGNC ID
|
HGNC:7029
|
|
OMIM ID
|
164860 |
|
Summary
|
This gene encodes a member of the receptor tyrosine kinase family of proteins and the product of the proto-oncogene MET. The encoded preproprotein is proteolytically processed to generate alpha and beta subunits that are linked via disulfide bonds to form the mature receptor. Further processing of the beta subunit results in the formation of the M10 peptide, which has been shown to reduce lung fibrosis. Binding of its ligand, hepatocyte growth factor, induces dimerization and activation of the receptor, which plays a role in cellular survival, embryogenesis, and cellular migration and invasion. Mutations in this gene are associated with papillary renal cell carcinoma, hepatocellular carcinoma, and various head and neck cancers. Amplification and overexpression of this gene are also associated with multiple human cancers. [provided by RefSeq, May 2016] |
Target gene [MET] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5016285 |
chr7 |
12 |
10 |
2 |
24 |
View |
| 5016286 |
chr7 |
3 |
2 |
0 |
5 |
View |
Target gene [MET] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE199029
|
Variation;RNA-seq |
44 |
NanoString Human nCounter PanCancer IO360 Panel |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE138080 - MET expression across samples
|
Volcano Plot
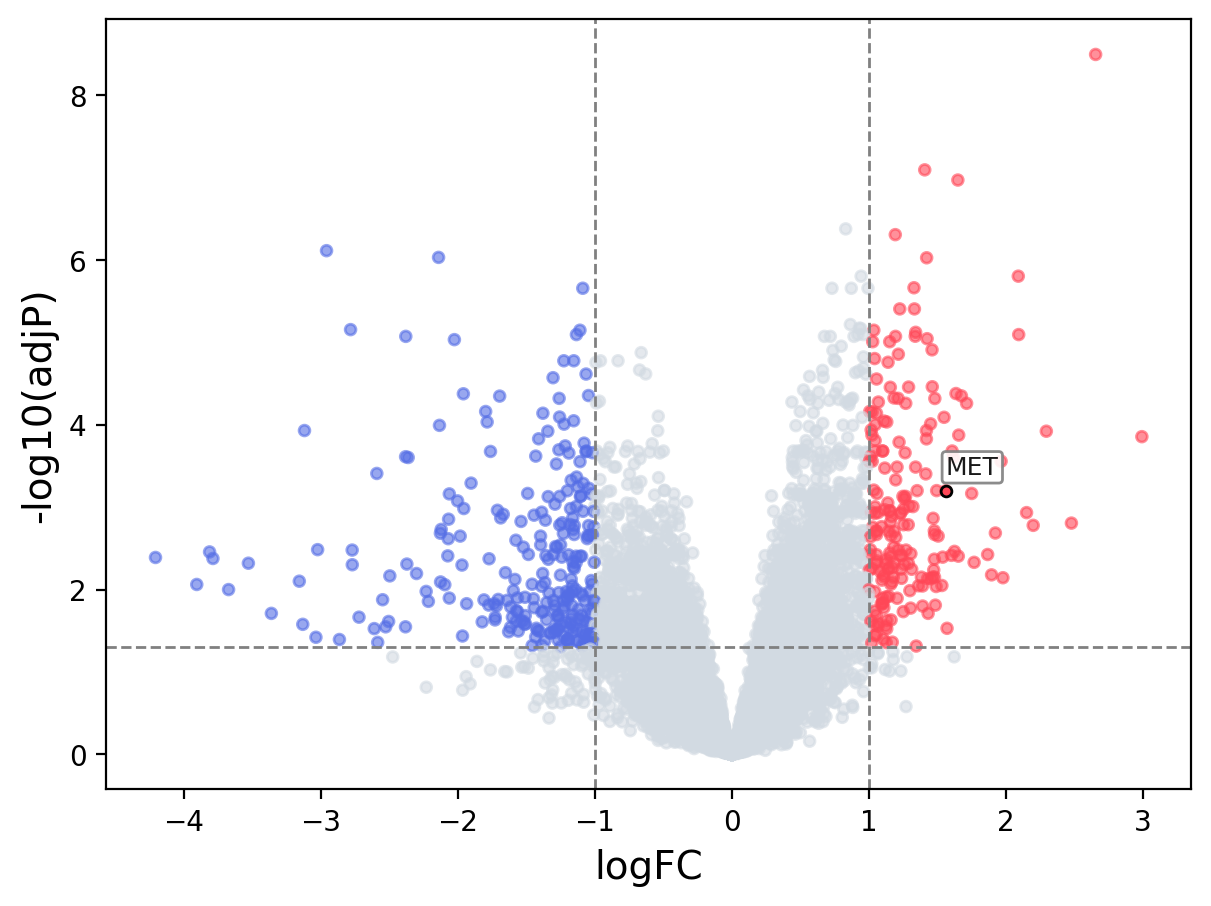
|
Bar Plot
|
> Dataset: TCGA_CESC - MET expression across samples
|
Volcano Plot
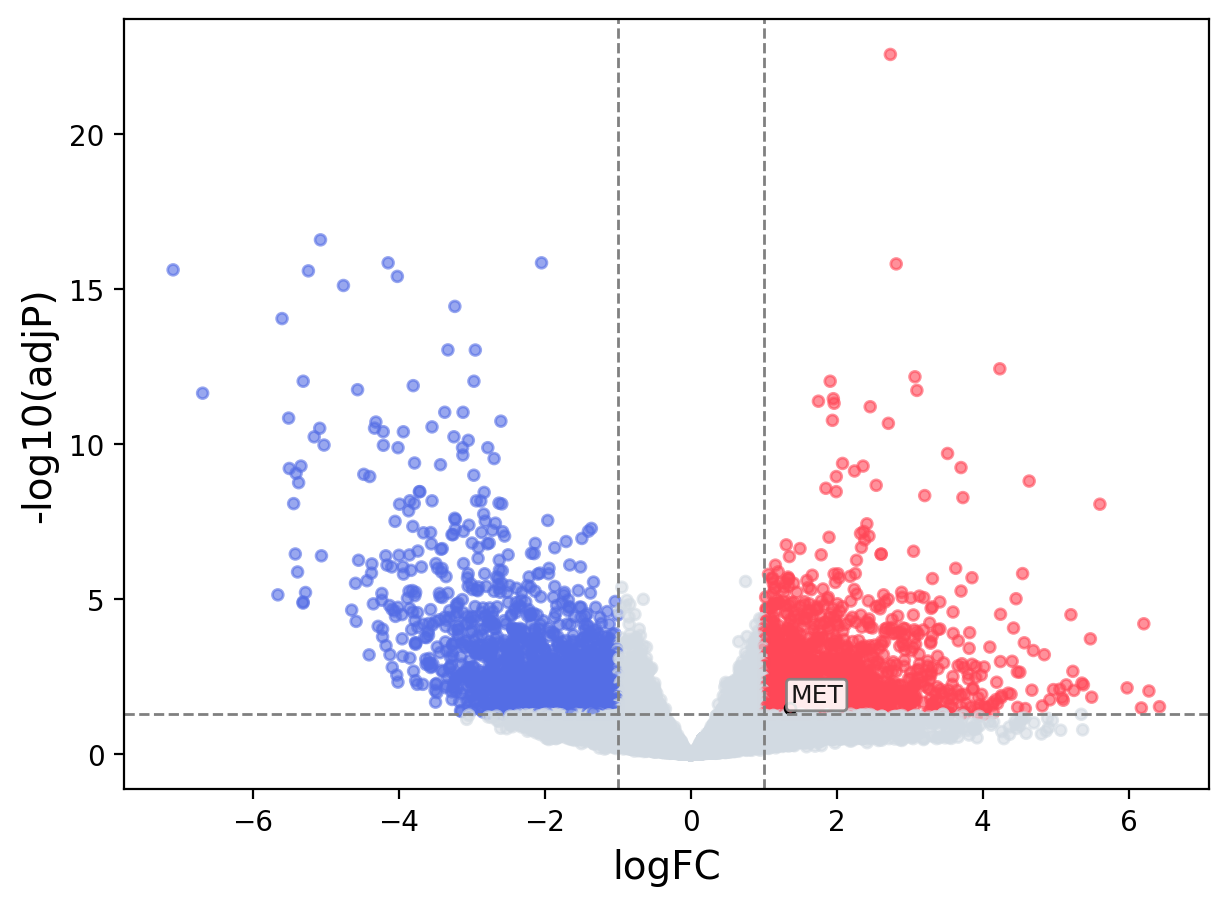
|
Bar Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information