Gene Information
|
Gene Name
|
FAS |
|
Gene ID
|
355
|
|
Gene Full Name
|
Fas cell surface death receptor |
|
Gene Alias
|
ALPS1A|APO-1|APT1|CD95|FAS1|FASTM|TNFRSF6 |
|
Transcripts
|
ENSG00000026103
|
|
Virus
|
EBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Nuclear bodies, Cytosol;Secreted to blood; 
|
|
Membrane Info
|
Cancer-related genes, Candidate cardiovascular disease genes, CD markers, Disease related genes, Human disease related genes, Predicted intracellular proteins, Predicted membrane proteins, Predicted secreted proteins |
|
Uniport_ID
|
P25445
|
|
HGNC ID
|
HGNC:11920
|
|
OMIM ID
|
134637 |
|
Summary
|
The protein encoded by this gene is a member of the TNF-receptor superfamily. This receptor contains a death domain. It has been shown to play a central role in the physiological regulation of programmed cell death, and has been implicated in the pathogenesis of various malignancies and diseases of the immune system. The interaction of this receptor with its ligand allows the formation of a death-inducing signaling complex that includes Fas-associated death domain protein (FADD), caspase 8, and caspase 10. The autoproteolytic processing of the caspases in the complex triggers a downstream caspase cascade, and leads to apoptosis. This receptor has been also shown to activate NF-kappaB, MAPK3/ERK1, and MAPK8/JNK, and is found to be involved in transducing the proliferating signals in normal diploid fibroblast and T cells. Several alternatively spliced transcript variants have been described, some of which are candidates for nonsense-mediated mRNA decay (NMD). The isoforms lacking the transmembrane domain may negatively regulate the apoptosis mediated by the full length isoform. [provided by RefSeq, Mar 2011] |
Target gene [FAS] related to virus gene/protein/region 
Mutation Table: if previous studies reported that target gene was related to virus gene/region/protein, the information in this literature was listed in this section
Target gene [FAS] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE42867
|
Expression array |
12 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
C GSE246060
|
Chip-seq |
26 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE45919
|
Expression array |
24 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE198332
|
Methylation profiling (Array) |
7 |
Infinium MethylationEPIC |
|
GSE223328
|
Expression array |
16 |
Agilent-072363 SurePrint G3 Human GE v3 8x60K Microarray 039494 [Probe Name Version] |
|
GSE208281
|
RNA-seq |
50 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE100458
|
Expression array |
14 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array [CDF: Brainarray HGU133Plus2_Hs_ENTREZG_v21] |
|
C GSE198334
|
Chip-seq |
4 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE165194
|
RNA-seq |
3 |
Illumina NextSeq 500 (Homo sapiens) |
|
C GSE246059
|
ATAC-seq |
8 |
Illumina NextSeq 500 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE177046
|
RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE135644
|
Expression array |
10 |
Sentrix Human-6 v2 Expression BeadChip |
|
C GSE225248
|
Chip-seq |
12 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE102203
|
Expression array |
10 |
[HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array [probe set (exon) version] |
|
GSE85599
|
Expression array |
17 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE12452
|
Expression array |
41 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
GSE58240
|
Expression array |
32 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181906
|
RNA-seq |
24 |
Illumina NovaSeq 6000 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE246060 - FAS peak across samples
|
Peak Plot
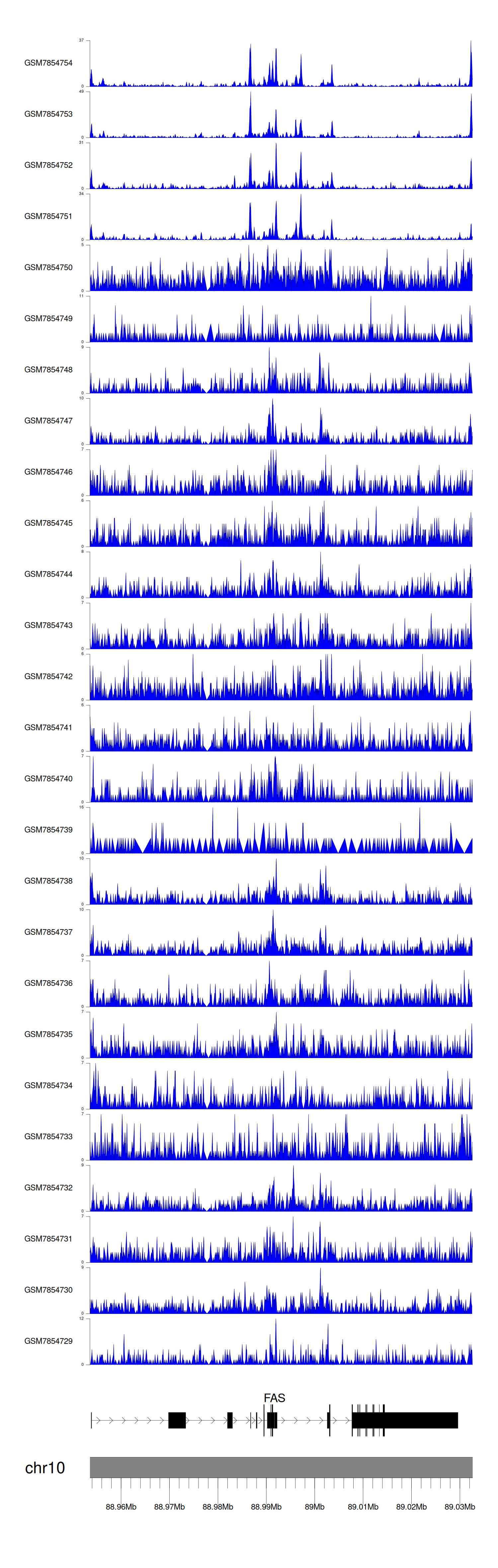
|
> Dataset: GSE198334 - FAS peak across samples
|
Peak Plot
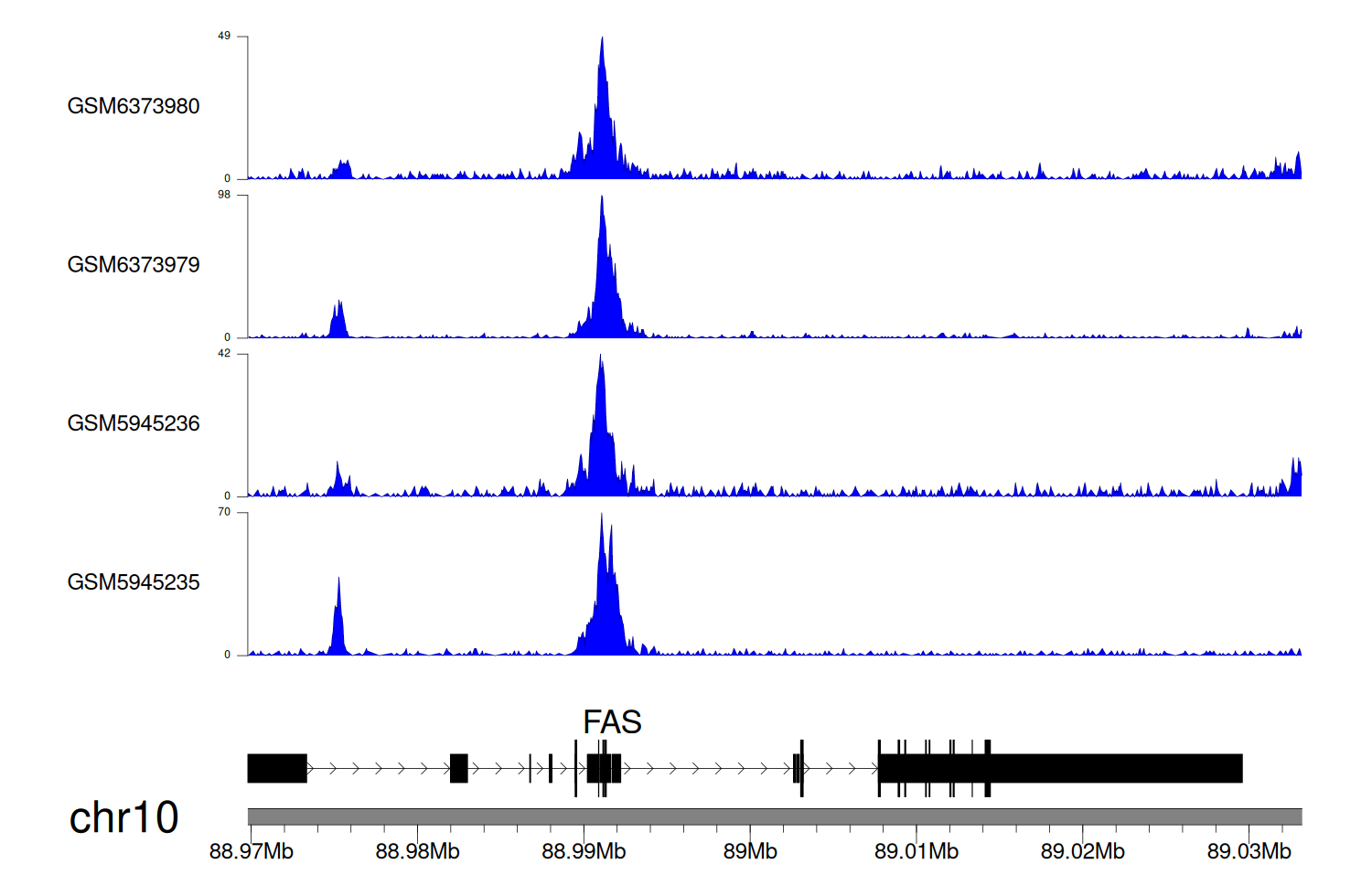
|
> Dataset: GSE246059 - FAS peak across samples
|
Peak Plot
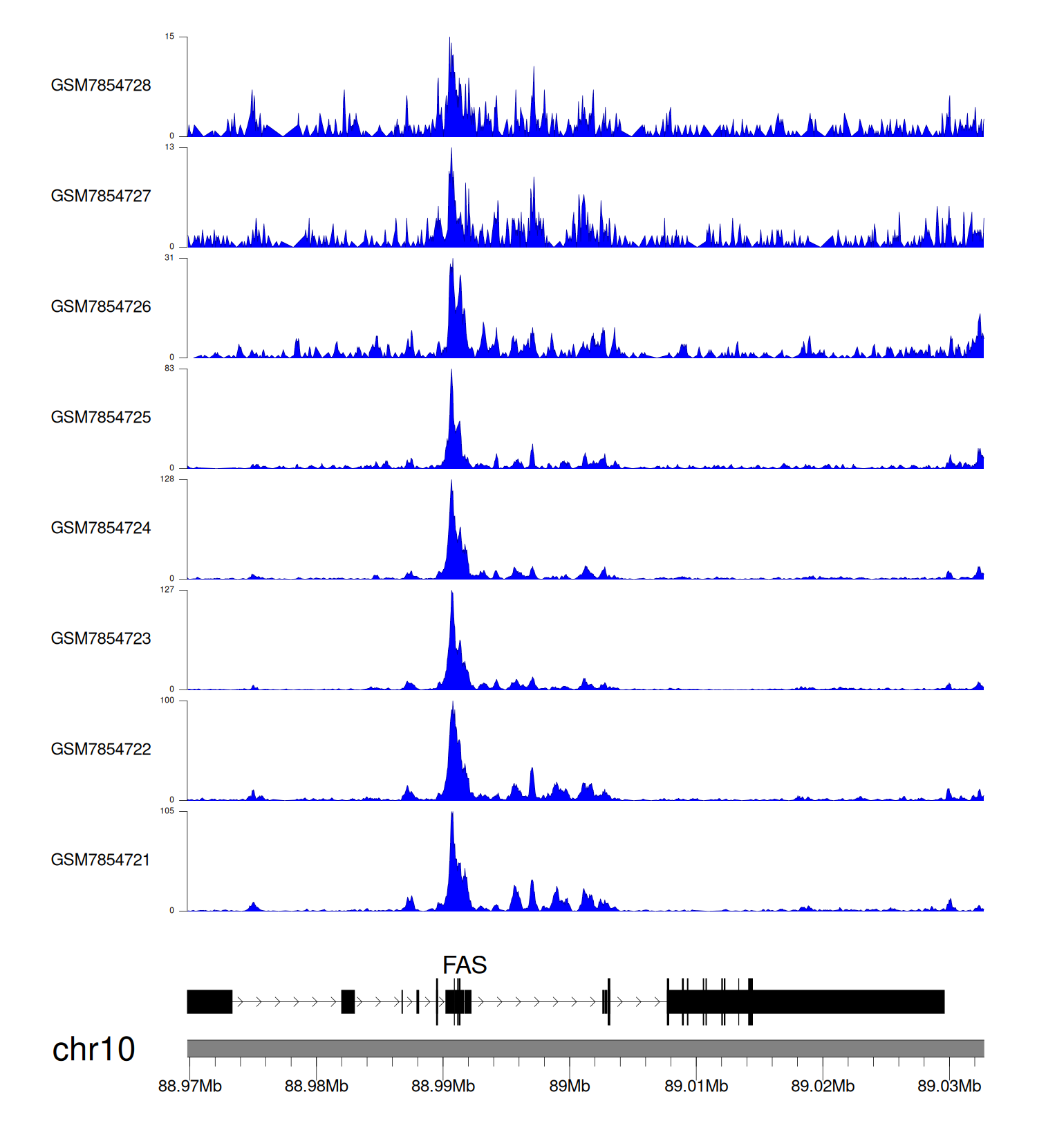
|
> Dataset: GSE225248 - FAS peak across samples
|
Peak Plot
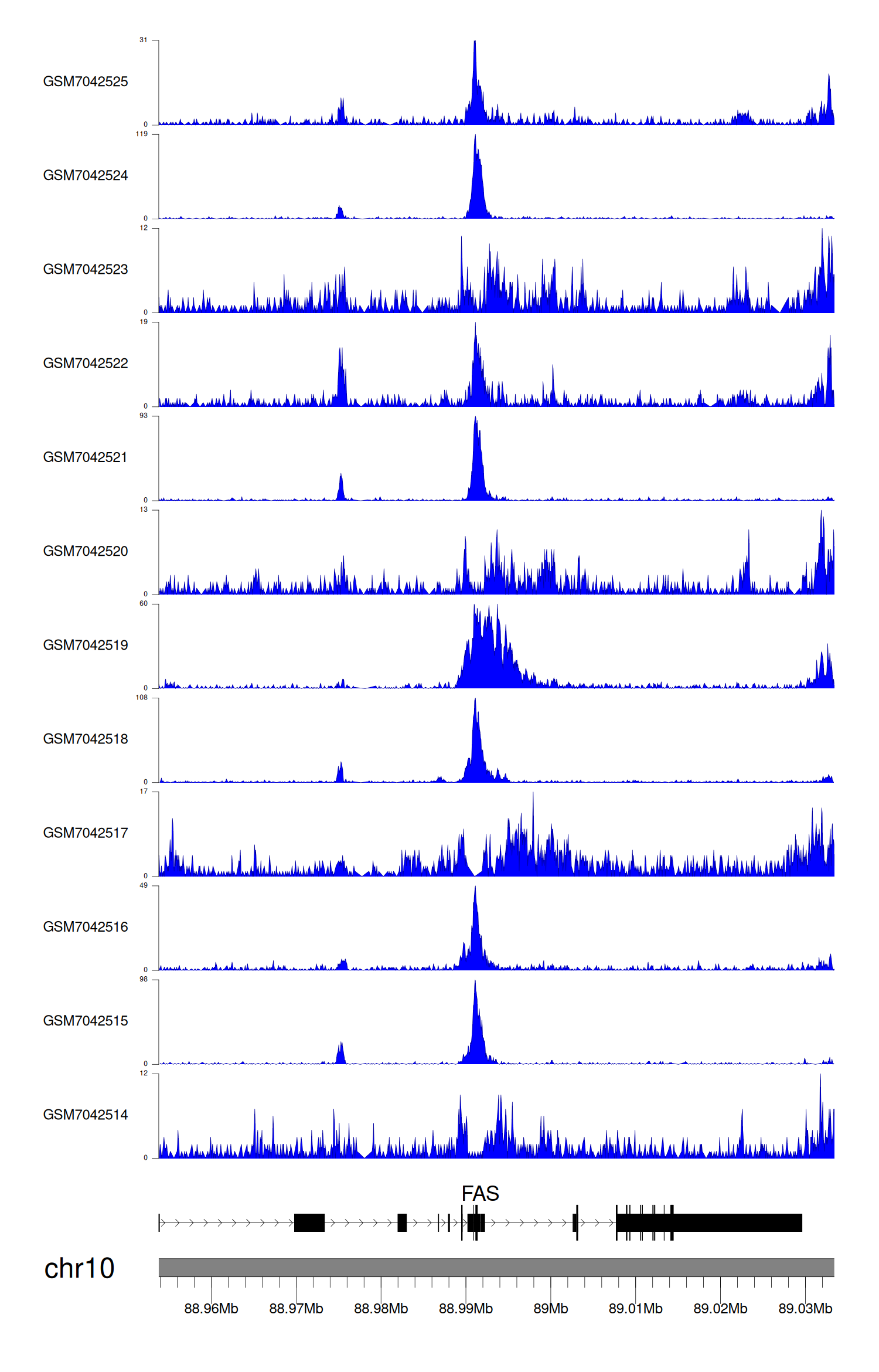
|
|
|
 EBV Target gene Detail Information
EBV Target gene Detail Information