Gene Information
|
Gene Name
|
PLEC |
|
Gene ID
|
5339
|
|
Gene Full Name
|
plectin |
|
Gene Alias
|
EBS1|EBS5A|EBS5B|EBS5C|EBS5D|EBSMD|EBSND|EBSO|EBSOG|EBSPA|HD1|LGMD2Q|LGMDR17|PCN|PLEC1|PLEC1b|PLTN |
|
Transcripts
|
ENSG00000178209
|
|
Virus
|
EBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Intermediate filaments, Cytosol;Focal adhesion sites; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins |
|
Uniport_ID
|
Q15149
|
|
HGNC ID
|
HGNC:9069
|
|
OMIM ID
|
601282 |
|
Summary
|
Plectin is a prominent member of an important family of structurally and in part functionally related proteins, termed plakins or cytolinkers, that are capable of interlinking different elements of the cytoskeleton. Plakins, with their multi-domain structure and enormous size, not only play crucial roles in maintaining cell and tissue integrity and orchestrating dynamic changes in cytoarchitecture and cell shape, but also serve as scaffolding platforms for the assembly, positioning, and regulation of signaling complexes (reviewed in PMID: 9701547, 11854008, and 17499243). Plectin is expressed as several protein isoforms in a wide range of cell types and tissues from a single gene located on chromosome 8 in humans (PMID: 8633055, 8698233). Until 2010, this locus was named plectin 1 (symbol PLEC1 in human; Plec1 in mouse and rat) and the gene product had been referred to as "hemidesmosomal protein 1" or "plectin 1, intermediate filament binding 500kDa". These names were superseded by plectin. The plectin gene locus in mouse on chromosome 15 has been analyzed in detail (PMID: 10556294, 14559777), revealing a genomic exon-intron organization with well over 40 exons spanning over 62 kb and an unusual 5' transcript complexity of plectin isoforms. Eleven exons (1-1j) have been identified that alternatively splice directly into a common exon 2 which is the first exon to encode plectin's highly conserved actin binding domain (ABD). Three additional exons (-1, 0a, and 0) splice into an alternative first coding exon (1c), and two additional exons (2alpha and 3alpha) are optionally spliced within the exons encoding the acting binding domain (exons 2-8). Analysis of the human locus has identified eight of the eleven alternative 5' exons found in mouse and rat (PMID: 14672974); exons 1i, 1j and 1h have not been confirmed in human. Furthermore, isoforms lacking the central rod domain encoded by exon 31 have been detected in mouse (PMID:10556294), rat (PMID: 9177781), and human (PMID: 11441066, 10780662, 20052759). The short alternative amino-terminal sequences encoded by the different first exons direct the targeting of the various isoforms to distinct subcellular locations (PMID: 14559777). As the expression of specific plectin isoforms was found to be dependent on cell type (tissue) and stage of development (PMID: 10556294, 12542521, 17389230) it appears that each cell type (tissue) contains a unique set (proportion and composition) of plectin isoforms, as if custom-made for specific requirements of the particular cells. Concordantly, individual isoforms were found to carry out distinct and specific functions (PMID: 14559777, 12542521, 18541706). In 1996, a number of groups reported that patients suffering from epidermolysis bullosa simplex with muscular dystrophy (EBS-MD) lacked plectin expression in skin and muscle tissues due to defects in the plectin gene (PMID: 8698233, 8941634, 8636409, 8894687, 8696340). Two other subtypes of plectin-related EBS have been described: EBS-pyloric atresia (PA) and EBS-Ogna. For reviews of plectin-related diseases see PMID: 15810881, 19945614. Mutations in the plectin gene related to human diseases should be named based on the position in NM_000445 (variant 1, isoform 1c), unless the mutation is located within one of the other alternative first exons, in which case the position in the respective Reference Sequence should be used. [provided by RefSeq, Aug 2011] |
Target gene [PLEC] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 3001242 |
chr8 |
17 |
6 |
0 |
23 |
View |
Target gene [PLEC] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE42867
|
Expression array |
12 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
C GSE246060
|
Chip-seq |
26 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE45919
|
Expression array |
24 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
C GSE198334
|
Chip-seq |
4 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE223328
|
Expression array |
16 |
Agilent-072363 SurePrint G3 Human GE v3 8x60K Microarray 039494 [Probe Name Version] |
|
GSE208281
|
RNA-seq |
50 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE100458
|
Expression array |
14 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array [CDF: Brainarray HGU133Plus2_Hs_ENTREZG_v21] |
|
GSE165194
|
RNA-seq |
3 |
Illumina NextSeq 500 (Homo sapiens) |
|
C GSE246059
|
ATAC-seq |
8 |
Illumina NextSeq 500 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE177046
|
RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE225248
|
Chip-seq |
12 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE102203
|
Expression array |
10 |
[HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array [probe set (exon) version] |
|
GSE85599
|
Expression array |
17 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE12452
|
Expression array |
41 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
GSE58240
|
Expression array |
32 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181906
|
RNA-seq |
24 |
Illumina NovaSeq 6000 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE45919 - PLEC expression across samples
|
Volcano Plot
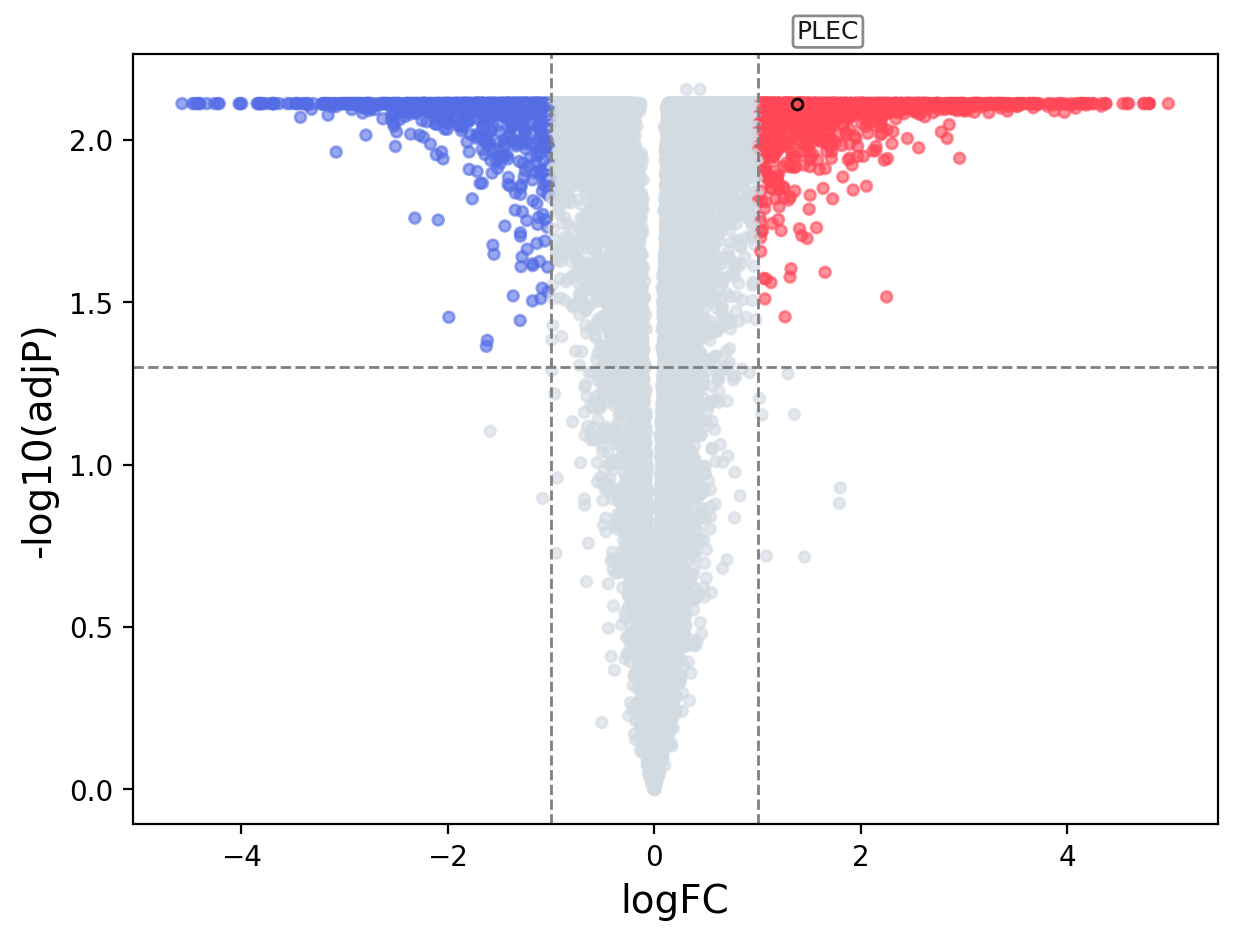
|
Bar Plot
|
|
|
 EBV Target gene Detail Information
EBV Target gene Detail Information