Gene Information
|
Gene Name
|
PPARA |
|
Gene ID
|
5465
|
|
Gene Full Name
|
peroxisome proliferator activated receptor alpha |
|
Gene Alias
|
NR1C1|PPAR|PPAR-alpha|PPARalpha|hPPAR |
|
Transcripts
|
ENSG00000186951
|
|
Virus
|
EBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Primary cilium, Centriolar satellite, Basal body; 
|
|
Membrane Info
|
FDA approved drug targets, Metabolic proteins, Nuclear receptors, Predicted intracellular proteins, Transcription factors, Transporters |
|
Uniport_ID
|
Q07869
|
|
HGNC ID
|
HGNC:9232
|
|
OMIM ID
|
170998 |
|
Summary
|
Peroxisome proliferators include hypolipidemic drugs, herbicides, leukotriene antagonists, and plasticizers; this term arises because they induce an increase in the size and number of peroxisomes. Peroxisomes are subcellular organelles found in plants and animals that contain enzymes for respiration and for cholesterol and lipid metabolism. The action of peroxisome proliferators is thought to be mediated via specific receptors, called PPARs, which belong to the steroid hormone receptor superfamily. PPARs affect the expression of target genes involved in cell proliferation, cell differentiation and in immune and inflammation responses. Three closely related subtypes (alpha, beta/delta, and gamma) have been identified. This gene encodes the subtype PPAR-alpha, which is a nuclear transcription factor. Multiple alternatively spliced transcript variants have been described for this gene, although the full-length nature of only two has been determined. [provided by RefSeq, Jul 2008] |
Target gene [PPARA] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 3000704 |
chr22 |
4 |
2 |
0 |
6 |
View |
Target gene [PPARA] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE42867
|
Expression array |
12 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
C GSE246060
|
Chip-seq |
26 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE45919
|
Expression array |
24 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE198332
|
Methylation profiling (Array) |
7 |
Infinium MethylationEPIC |
|
GSE223328
|
Expression array |
16 |
Agilent-072363 SurePrint G3 Human GE v3 8x60K Microarray 039494 [Probe Name Version] |
|
GSE208281
|
RNA-seq |
50 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE100458
|
Expression array |
14 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array [CDF: Brainarray HGU133Plus2_Hs_ENTREZG_v21] |
|
C GSE198334
|
Chip-seq |
4 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE165194
|
RNA-seq |
3 |
Illumina NextSeq 500 (Homo sapiens) |
|
C GSE246059
|
ATAC-seq |
8 |
Illumina NextSeq 500 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE177046
|
RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE135644
|
Expression array |
10 |
Sentrix Human-6 v2 Expression BeadChip |
|
C GSE225248
|
Chip-seq |
12 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE102203
|
Expression array |
10 |
[HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array [probe set (exon) version] |
|
GSE85599
|
Expression array |
17 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE12452
|
Expression array |
41 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
GSE58240
|
Expression array |
32 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181906
|
RNA-seq |
24 |
Illumina NovaSeq 6000 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE246060 - PPARA peak across samples
|
Peak Plot
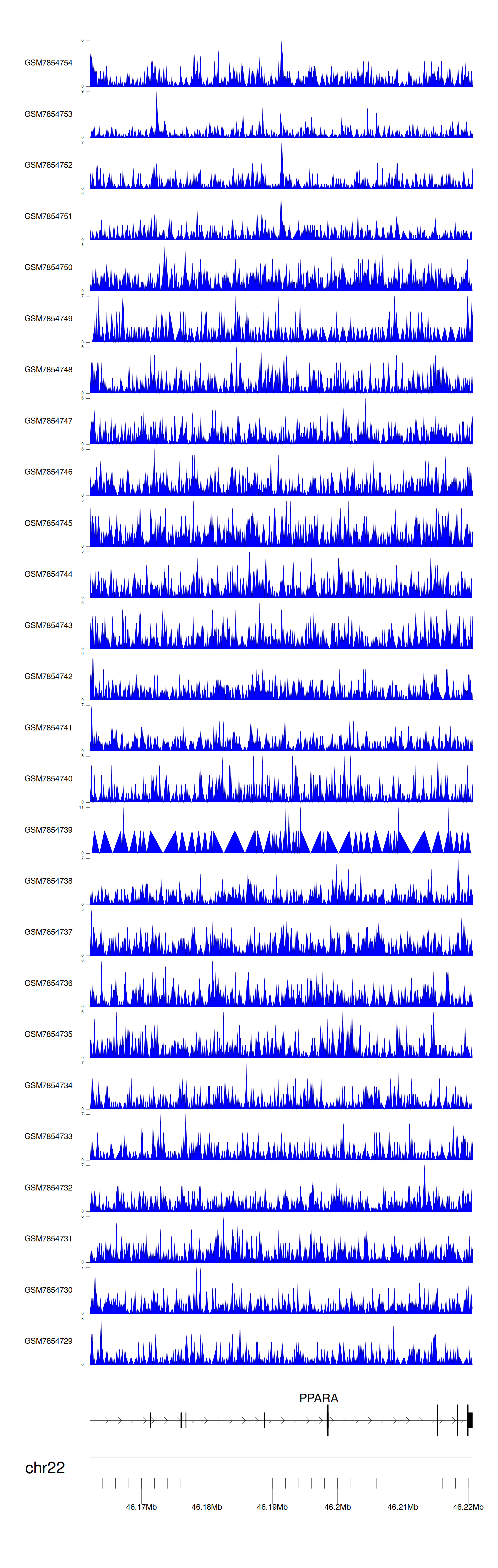
|
> Dataset: GSE198334 - PPARA peak across samples
|
Peak Plot
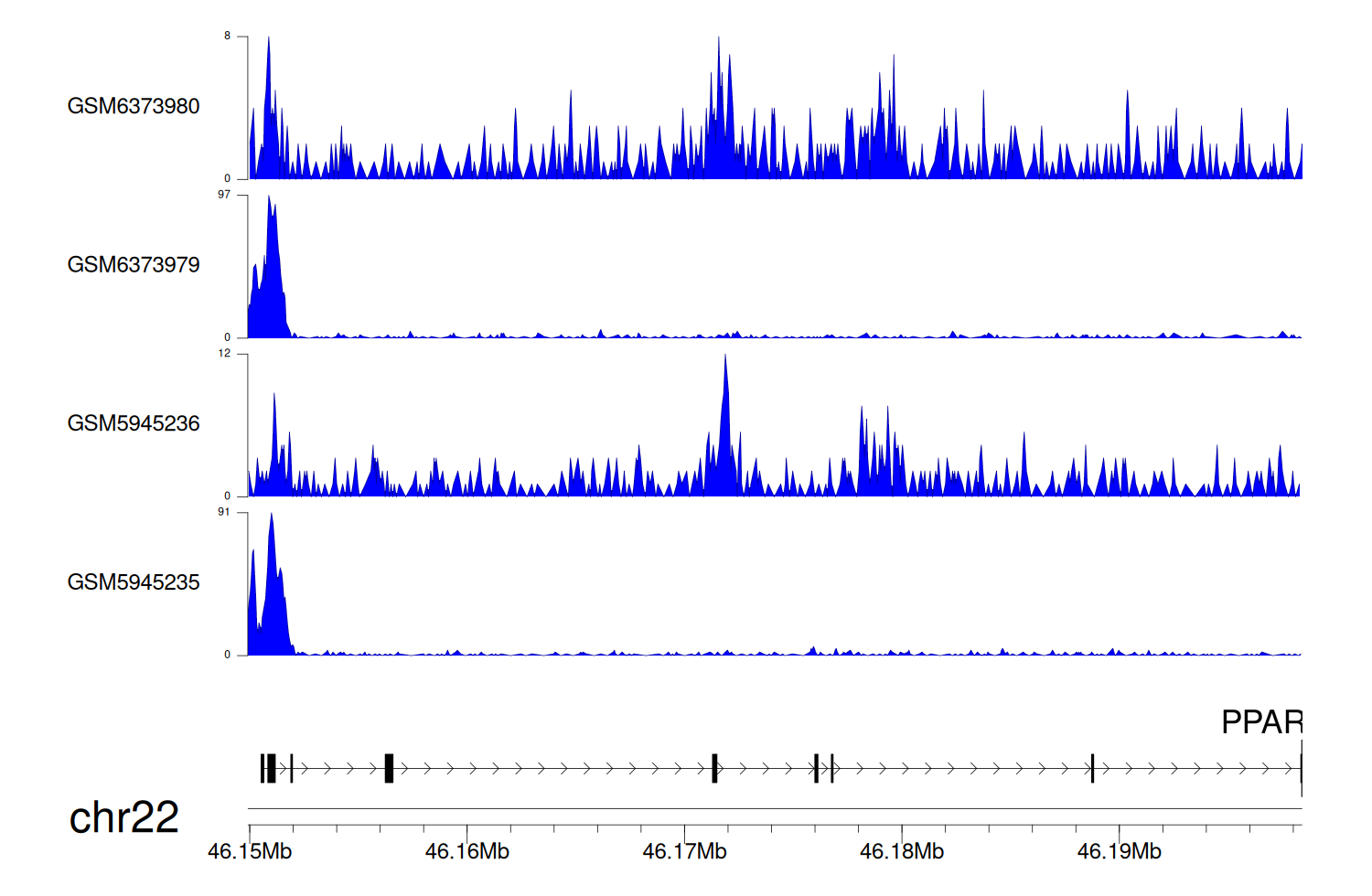
|
> Dataset: GSE246059 - PPARA peak across samples
|
Peak Plot
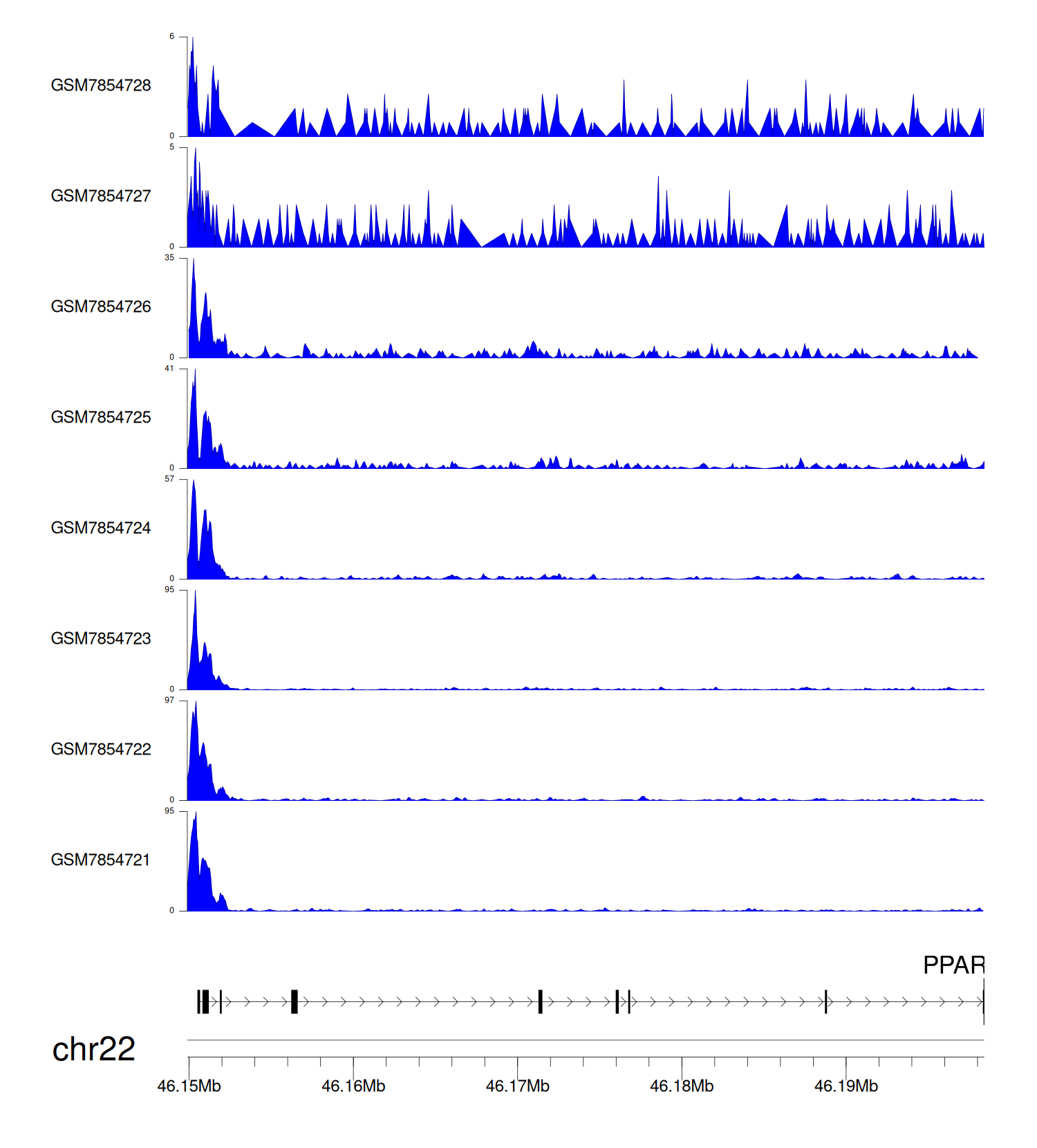
|
> Dataset: GSE225248 - PPARA peak across samples
|
Peak Plot
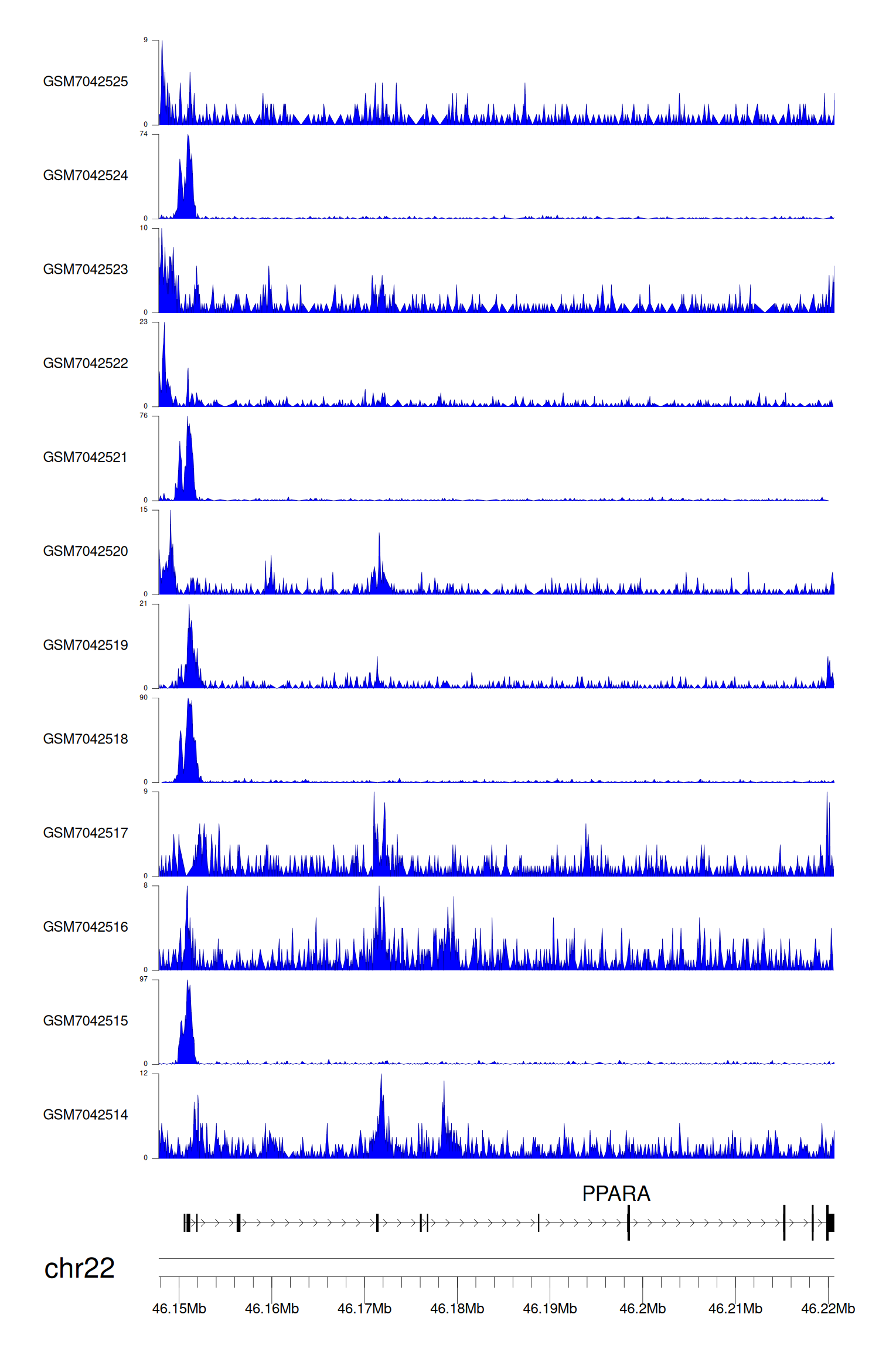
|
|
|
 EBV Target gene Detail Information
EBV Target gene Detail Information