Gene Information
|
Gene Name
|
S100B |
|
Gene ID
|
6285
|
|
Gene Full Name
|
S100 calcium binding protein B |
|
Gene Alias
|
NEF|S100|S100-B|S100beta |
|
Transcripts
|
ENSG00000160307
|
|
Virus
|
EBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Primary cilium, Cytosol;Nucleoplasm, Vesicles, Microtubules, Basal body;Secreted - unknown location; 
|
|
Membrane Info
|
Cancer-related genes, Candidate cardiovascular disease genes, Predicted intracellular proteins, Transporters |
|
Uniport_ID
|
P04271
|
|
HGNC ID
|
HGNC:10500
|
|
OMIM ID
|
176990 |
|
Summary
|
The protein encoded by this gene is a member of the S100 family of proteins containing 2 EF-hand calcium-binding motifs. S100 proteins are localized in the cytoplasm and/or nucleus of a wide range of cells, and involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation. S100 genes include at least 13 members which are located as a cluster on chromosome 1q21; however, this gene is located at 21q22.3. This protein may function in Neurite extension, proliferation of melanoma cells, stimulation of Ca2+ fluxes, inhibition of PKC-mediated phosphorylation, astrocytosis and axonal proliferation, and inhibition of microtubule assembly. Chromosomal rearrangements and altered expression of this gene have been implicated in several neurological, neoplastic, and other types of diseases, including Alzheimer's disease, Down's syndrome, epilepsy, amyotrophic lateral sclerosis, melanoma, and type I diabetes. [provided by RefSeq, Jul 2008] |
Target gene [S100B] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 3000053 |
chr21 |
68 |
151 |
286 |
505 |
View |
Target gene [S100B] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE42867
|
Expression array |
12 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
C GSE246060
|
Chip-seq |
26 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE45919
|
Expression array |
24 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE198332
|
Methylation profiling (Array) |
7 |
Infinium MethylationEPIC |
|
GSE223328
|
Expression array |
16 |
Agilent-072363 SurePrint G3 Human GE v3 8x60K Microarray 039494 [Probe Name Version] |
|
GSE208281
|
RNA-seq |
50 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE100458
|
Expression array |
14 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array [CDF: Brainarray HGU133Plus2_Hs_ENTREZG_v21] |
|
GSE165194
|
RNA-seq |
3 |
Illumina NextSeq 500 (Homo sapiens) |
|
C GSE246059
|
ATAC-seq |
8 |
Illumina NextSeq 500 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE177046
|
RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE135644
|
Expression array |
10 |
Sentrix Human-6 v2 Expression BeadChip |
|
C GSE225248
|
Chip-seq |
12 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE102203
|
Expression array |
10 |
[HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array [probe set (exon) version] |
|
GSE85599
|
Expression array |
17 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE12452
|
Expression array |
41 |
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
|
GSE58240
|
Expression array |
32 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
E GSE181906
|
RNA-seq |
24 |
Illumina NovaSeq 6000 (Homo sapiens) |
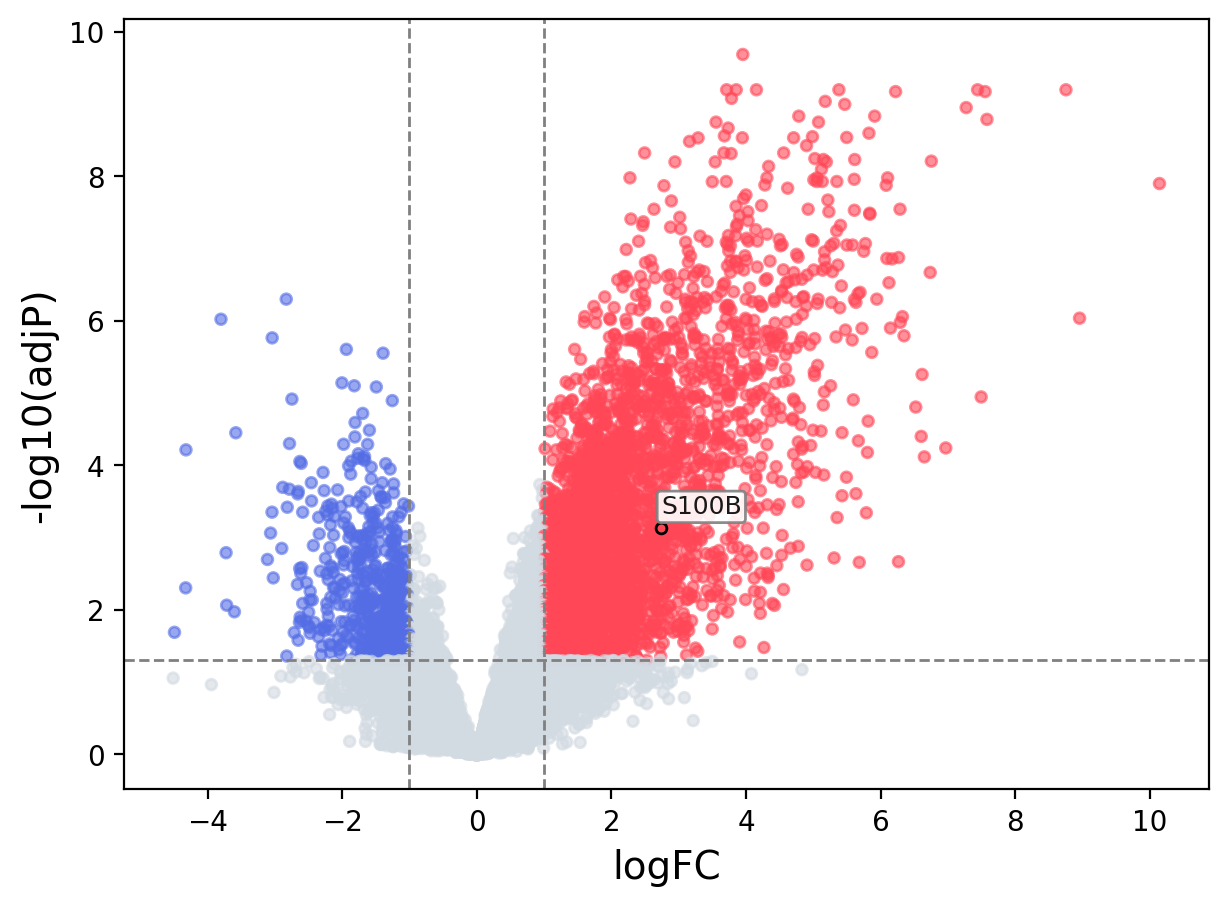
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE181906 - S100B expression across samples
|
Volcano Plot
|
Bar Plot
|
|
|
 EBV Target gene Detail Information
EBV Target gene Detail Information