Gene Information
|
Gene Name
|
FANCD2 |
|
Gene ID
|
2177
|
|
Gene Full Name
|
FA complementation group D2 |
|
Gene Alias
|
FA-D2|FA4|FACD|FAD|FAD2|FANCD |
|
Transcripts
|
ENSG00000144554
|
|
Virus
|
HIV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm, Nuclear bodies;Nucleoli, Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Predicted intracellular proteins |
|
Uniport_ID
|
Q9BXW9
|
|
HGNC ID
|
HGNC:3585
|
|
OMIM ID
|
613984 |
|
Summary
|
The Fanconi anemia complementation group (FANC) currently includes FANCA, FANCB, FANCC, FANCD1 (also called BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ (also called BRIP1), FANCL, FANCM and FANCN (also called PALB2). The previously defined group FANCH is the same as FANCA. Fanconi anemia is a genetically heterogeneous recessive disorder characterized by cytogenetic instability, hypersensitivity to DNA crosslinking agents, increased chromosomal breakage, and defective DNA repair. The members of the Fanconi anemia complementation group do not share sequence similarity; they are related by their assembly into a common nuclear protein complex. This gene encodes the protein for complementation group D2. This protein is monoubiquinated in response to DNA damage, resulting in its localization to nuclear foci with other proteins (BRCA1 AND BRCA2) involved in homology-directed DNA repair. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016] |
Target gene [FANCD2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 4003637 |
chr3 |
48 |
9 |
0 |
57 |
View |
| 4012979 |
chr3 |
4 |
0 |
0 |
4 |
View |
Target gene [FANCD2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE99737
|
RNA-seq |
9 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE144332
|
RNA-seq |
75 |
NextSeq 550 (Homo sapiens) |
|
GSE164264
|
RNA-seq |
6 |
BGISEQ-500 (Homo sapiens) |
|
C GSE30738
|
Chip-seq |
2 |
Illumina Genome Analyzer II (Homo sapiens) |
|
GSE262621
|
RNA-seq |
70 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE128121
|
RNA-seq |
16 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE162914
|
RNA-seq |
211 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE187515
|
scRNA-seq |
104 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 4000 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE279982
|
Methylation profiling (Array) |
576 |
Infinium MethylationEPIC |
|
C GSE144329
|
ATAC-seq |
96 |
NextSeq 550 (Homo sapiens) |
|
GSE205204
|
RNA-seq |
14 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE125686
|
RNA-seq |
62 |
Illumina HiSeq 2000 (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens);Illumina NextSeq 500 (Homo sapiens) |
|
GSE198339
|
scRNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE233747
|
scRNA-seq |
40 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE165708
|
RNA-seq |
100 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE211651
|
Chip-seq;RNA-seq;PRO-seq |
39 |
Illumina NextSeq 500 (Homo sapiens);Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100376
|
ATAC-seq |
24 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE167211
|
RNA-seq |
1 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE165132
|
RNA-seq |
56 |
NextSeq 550 (Homo sapiens) |
|
C GSE100266
|
ATAC-seq;Chip-seq;RNA-seq |
32 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE246082
|
RNA-seq |
6 |
Illumina HiSeq 4000 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE30738 - FANCD2 peak across samples
|
Peak Plot
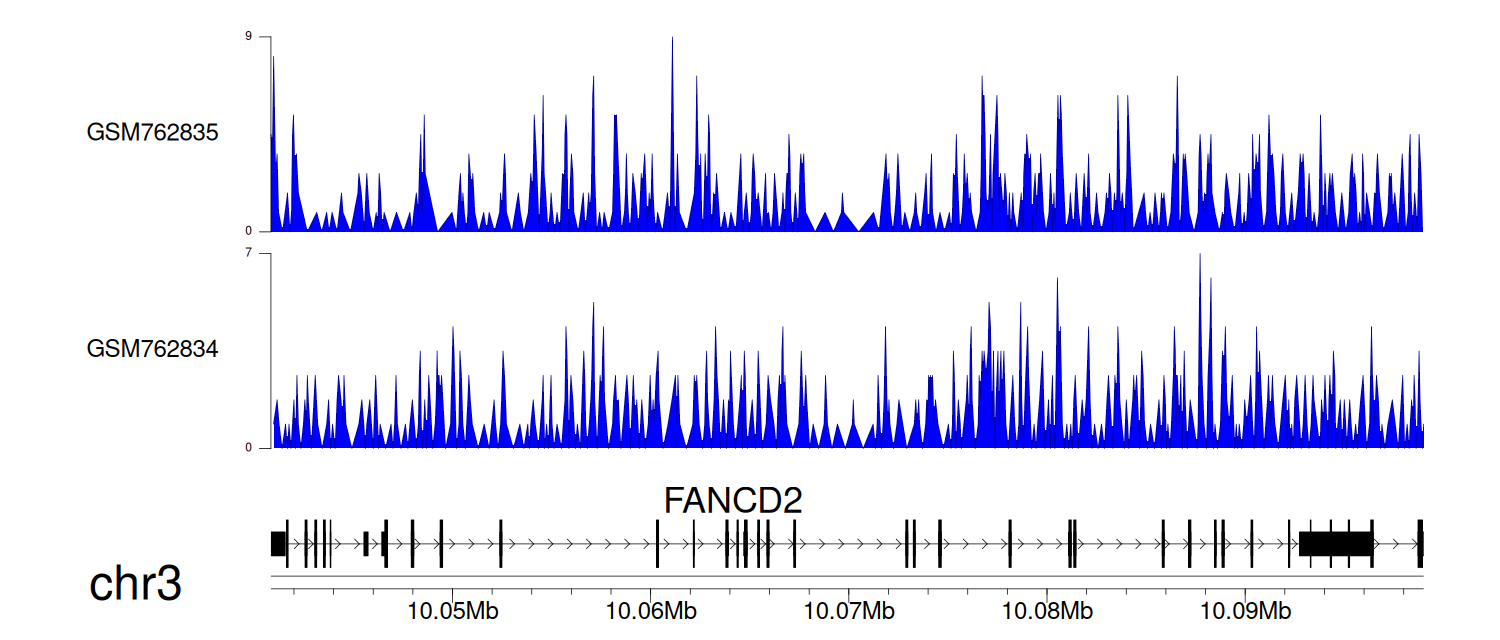
|
> Dataset: GSE144329 - FANCD2 peak across samples
|
Peak Plot

|
> Dataset: GSE100376 - FANCD2 peak across samples
|
Peak Plot
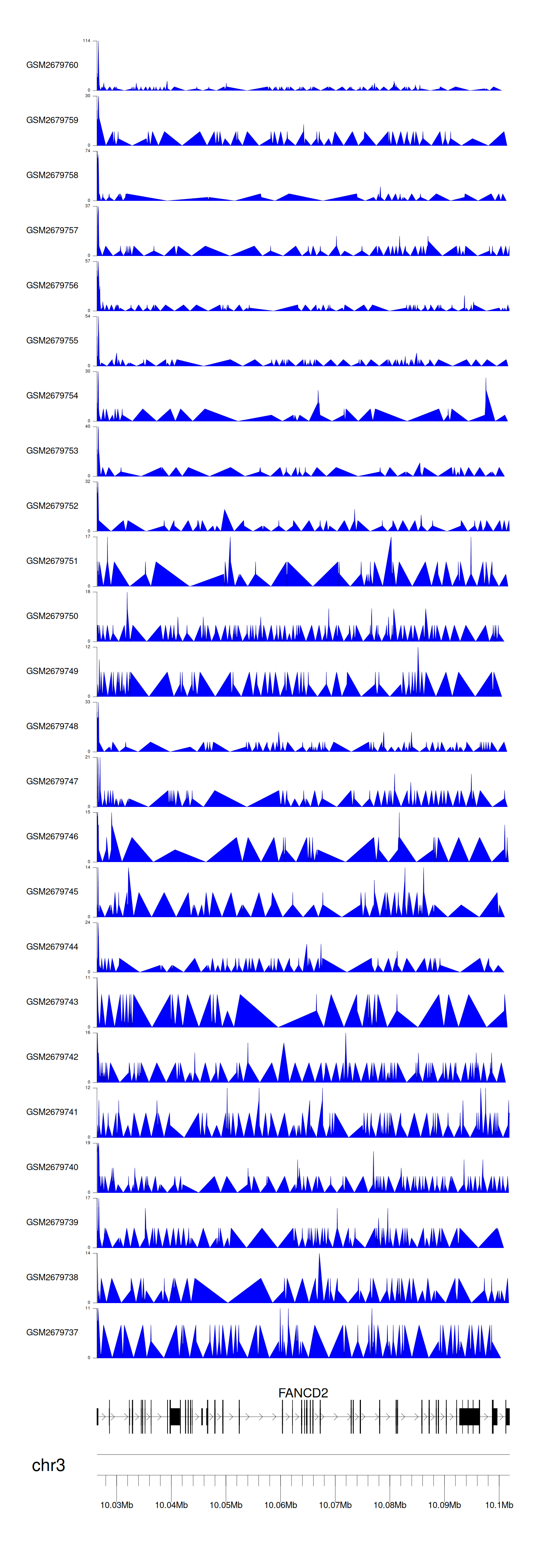
|
> Dataset: GSE100266 - FANCD2 peak across samples
|
Peak Plot
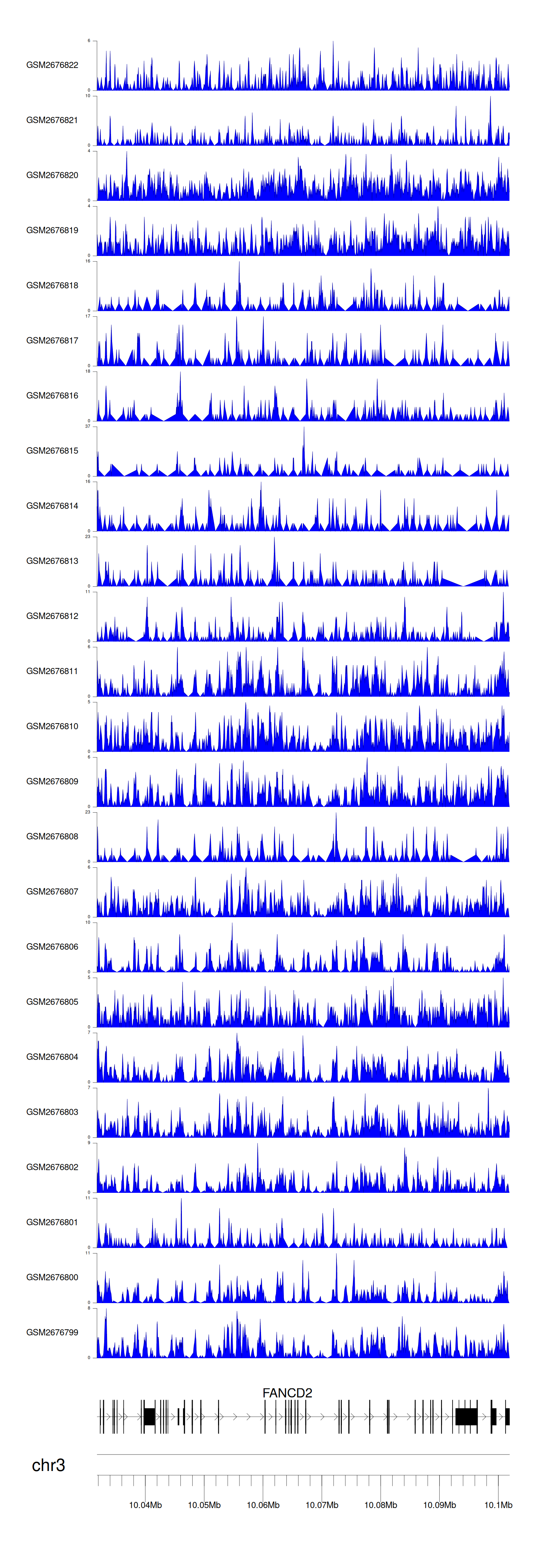
|
|
|
 HIV Target gene Detail Information
HIV Target gene Detail Information