| Accession |
GSE176428
|
| Status |
2021/6/10 |
| Title |
Dengue and Zika RNA-RNA interactomes reveal virus permissive and restrictive factors in human cells |
| Organism |
Homo sapiens |
| Experiment type |
Expression profiling by high throughput sequencing |
| Summary |
Identifying host factors is key to understanding RNA virus pathogenicity. Besides proteins, RNAs can interact with virus genomes to impact replication. Here, we used proximity ligation sequencing to identify virus-host RNA interactions for four strains of Zika virus (ZIKV) and one strain of dengue virus (DENV-1) in human cells. We found hundreds of coding and non-coding RNAs that bind to DENV and ZIKV viruses. Host RNAs tend to bind to single-stranded regions along the virus genomes and the binding is primarily driven by hybridization energetics. We observed that virus interacting host RNAs tend to be downregulated upon virus infection and identified a conserved set of virus responders that binds to most DENV and ZIKV. Knockdown of several short non-coding RNAs, including miR19a-3p, SCARNA2 and 7SK RNA resulted in a decrease in virus growth, suggesting that they act as virus permissive factors. In addition, the 3’UTR of DYNLT1 mRNA acts as a virus restrictive factor by binding to the conserved dumbbell region on DENV and ZIKV 3’UTR to decrease virus replication. This study demonstrates that host RNAs in themselves can impact virus growth in permissive and restrictive ways, expanding our understanding of host factors and RNA-based gene regulation during virus pathogenesis. |
| Samples |
| GSM ID |
Sample info |
Characteristics |
Description |
|
GSM5365352
|
hNPC 24hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:non-infected 24hr |
sample_4.rep |
|
GSM5365353
|
ZIKV+hNPC 0hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:ZIKV infected 0hr |
sample_5.rep |
|
GSM5365354
|
ZIKV+hNPC 2hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:ZIKV infected 2hr |
sample_6.rep |
|
GSM5365355
|
ZIKV+hNPC 16hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:ZIKV infected 16hr |
sample_7.rep |
|
GSM5365356
|
ZIKV+hNPC 24hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:ZIKV infected 24hr |
sample_8.rep |
|
GSM5365349
|
hNPC 0hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:non-infected 0hr |
sample_1.rep |
|
GSM5365350
|
hNPC 2hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:non-infected 2hr |
sample_2.rep |
|
GSM5365351
|
hNPC 16hr |
cell type:Human neuronal precursor cells; cell status:Differentiated from hESC into hNPC; treatment group:non-infected 16hr |
sample_3.rep |
|
| Platform |
GPL15520 : Illumina MiSeq (Homo sapiens) |
| Literature |
|
| Download |
Download xCell Data Download Expression Analysis Data
|
|
xCell Plot
|
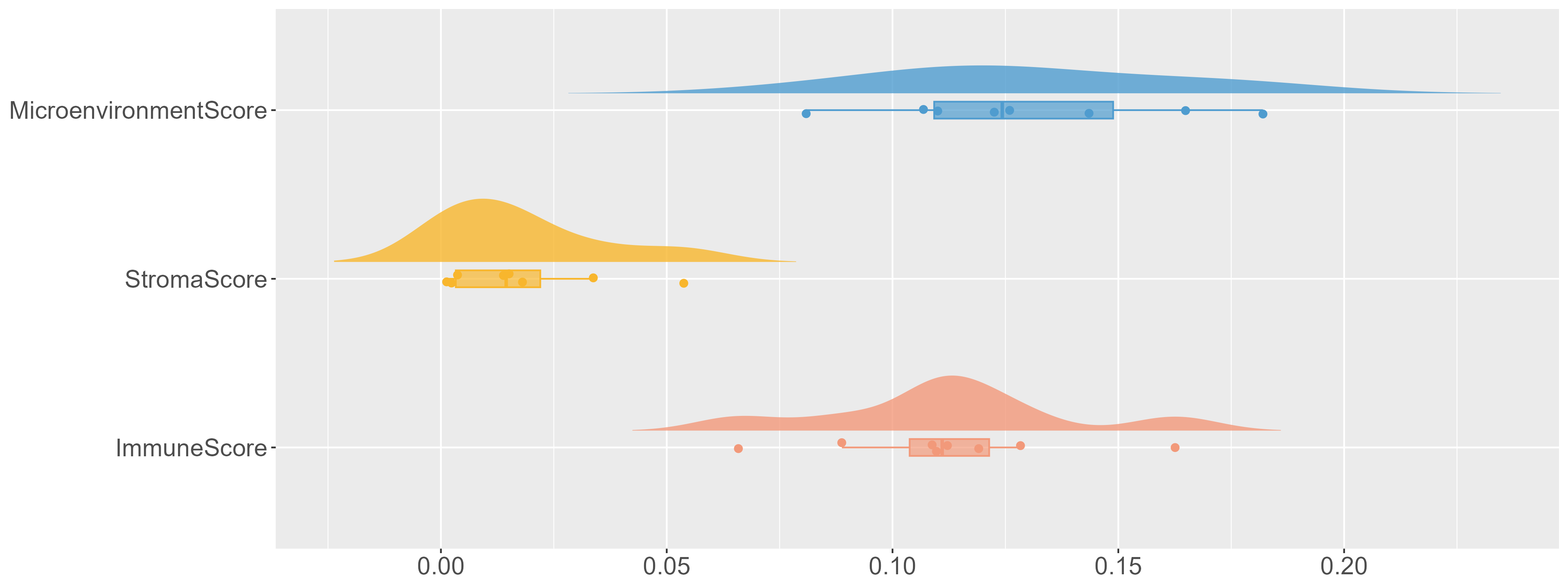
|
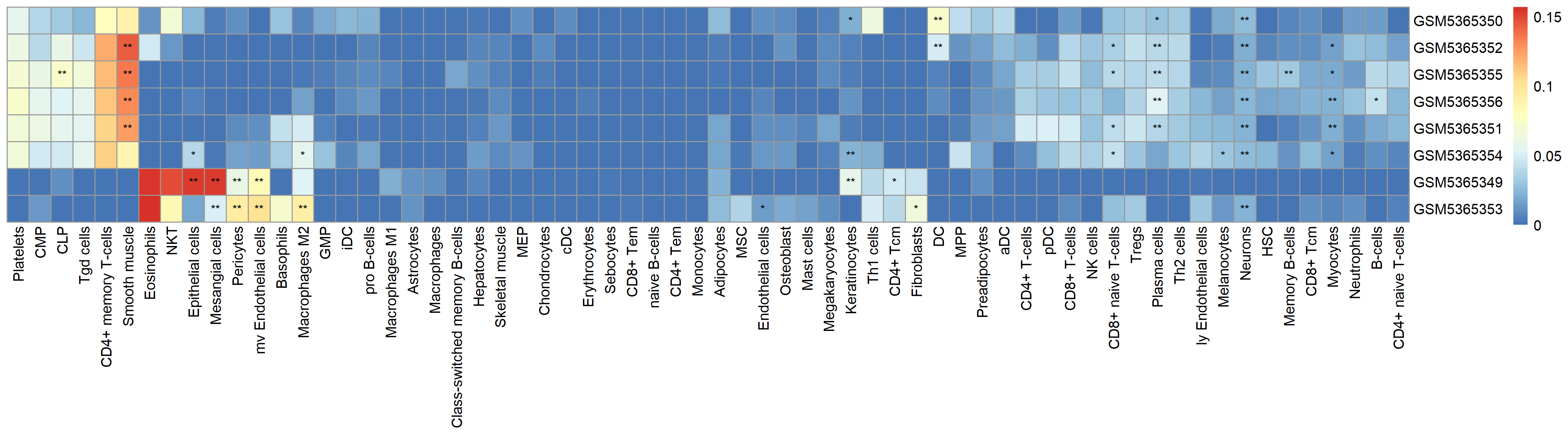 * p<0.05 ** p<0.01
* p<0.05 ** p<0.01
|
 Virus Dataset Detail
Virus Dataset Detail